Abstract
Hypertrophic cardiomyopathy (HCM) represents one of the most common heritable heart diseases. However, the signalling pathways and regulatory networks underlying the pathogenesis of HCM remain largely unknown. Here, we present a strand-specific RNA-seq dataset for both coding and lncRNA profiling in myocardial tissues from 28 HCM patients and 9 healthy donors. This dataset constitutes a valuable resource for the community to examine the dysregulated coding and lncRNA genes in HCM versus normal conditions.
Design Type(s) | transcription profiling design • disease state design • sequence analysis objective |
Measurement Type(s) | transcription profiling assay |
Technology Type(s) | RNA sequencing |
Factor Type(s) | experimental condition |
Sample Characteristic(s) | Homo sapiens • heart |
Machine-accessible metadata file describing the reported data (ISA-Tab format)
Similar content being viewed by others
Background & Summary
Hypertrophic cardiomyopathy (HCM) represents one of the most common heart diseases (an estimated prevalence of at least 0.2%), and a leading cause of sudden death in young people1,2. HCM is generally regarded as a genetic disorder caused predominately by mutations in eight sarcomere genes, including MYH7, MYBPC3, ACTC1, TPM1, MYL2, MYL3, TNNI3, and TNNT23. However, the signalling pathways and regulatory networks underlying the pathogenesis of HCM remain largely unknown.
Long non-coding RNAs (lncRNAs) are a large class of transcripts ≥200 nucleotides in length that do not encode proteins4. Compared with coding mRNAs, most lncRNAs are less well annotated and their functions are largely unexplored. Nevertheless, there is increasing evidence showing that lncRNAs are involved in a variety of biological processes and diseases5,6. lncRNAs have been implicated in pathologically processes of HCM, such as cardiomyocyte disarrangement, myocardial hypertrophy and interstitial fibrosis7. Through comparative analysis between 7 HCM patients and 5 control subjects using microarray, dysregulated lncRNAs in myocardial tissues of HCM patients were found to be involved in the pathogenesis of HCM through the regulation of pathogenetic pathways8.
RNA-seq is emerging as the major transcriptome profiling system. RNA-seq has considerable advantages over microarray in many aspects such as novel transcript identification through de novo assembly, splice junction identification and allele-specific expression analysis. Compared with the standard RNA-seq protocol, strand-specific RNA-seq retains strand of origin information, thus providing a greater resolution for sense/antisense profiling, which is essential for antisense lncRNA identification9. To our knowledge, we still lack a strand-specific RNA-seq dataset for myocardial tissues of HCM patients.
Here, we present a strand-specific RNA-seq dataset for both coding and lncRNA profiling in myocardial tissues from 28 HCM patients and 9 healthy donors. This dataset constitutes a valuable resource for the community to examine the dysregulated coding and lncRNA genes in HCM versus normal conditions. This dataset may also be reutilized through integration with future datasets to further enhance statistical power by increasing sample size.
Methods
Ethical approval
This study was approved by the ethics committee of the institutional review board at Fuwai Hospital. All procedures were conducted according to the ethical standards of the research committee. Informed consent was obtained from all subjects.
Patients and sample collection
Figure 1a shows the experimental design and workflow. We enrolled 28 HCM patients undergoing septal myectomy (Table 1 and Supplementary Table S1). All these patients were subjected to genetic testing to identify pathogenic mutations in HCM causal genes. Among them, 10 patients were confirmed to carry deleterious mutation in the gene MYBPC3 encoding cardiac myosin binding protein C, and 8 patients in the gene MYH7 encoding beta-myosin heavy chain. The remaining 10 patients were genetically undiagnosed. Myocardial tissues were collected during surgery and immediately placed in liquid nitrogen for storage. In addition, myocardial tissues in a normal, healthy condition (9 samples as a control group) were collected from donor hearts abandoned during cardiac transplant due to unexpected reasons.
Overview of the experimental procedure. (a) Schematic representation of the experimental workflow. The sampling position is indicated by a black rectangular. RNA isolation and library preparation for all samples were performed in the same batch. HCM: hypertrophic cardiomyopathy; GENETUN: Genetically undiagnosed HCM; MYBPC3: HCM patient with mutation in MYBPC3; MYH7: HCM patient with mutation in MYH7; NORMAL: Normal heart. (b) Bioinformatic analysis workflow.
RNA isolation and qualification
Total RNA was isolated with TRIzolTM reagent (Invitrogen, USA) according to the manufacturer’s instruction. RNA concentration was measured using Qubit® RNA Assay Kit in Qubit® 2.0 Fluorometer (Life Technologies, CA, USA). RNA purity was assessed using the NanoPhotometer® spectrophotometer (IMPLEN, CA, USA). RNA integrity was checked using the RNA Nano 6000 Assay Kit on the Agilent Bioanalyzer 2100 system (Agilent Technologies, CA, USA). Only samples with a 260:280 ratio of ≥1.5 and an RNA integrity number (RIN) of ≥8 were subjected to deep sequencing.
Strand-specific RNA-seq library preparation & sequencing
We prepared a strand-specific RNA-seq library for each sample. Firstly, ribosomal RNA (rRNA) was removed by Epicentre Ribo-zeroTM rRNA Removal Kit (Epicentre, USA) from 3 μg total RNA. Then, sequencing libraries were generated using NEBNext® UltraTM Directional RNA Library Prep Kit for Illumina® (NEB, USA) following manufacturer’s instructions. Briefly, the first strand cDNA synthesis was performed using M-MuLV reverse transcriptase and random hexamer primer. The second strand cDNA was synthesized using RNase H and DNA Polymerase I. The dTTP was replaced by dUTP in the reaction buffer. Following end repair and adenylation, cDNA fragments were ligated to adaptors. Then, 3 μl USER Enzyme was incubated with the cDNA for 15 min at 37 °C followed by 5 min at 95 °C before PCR. Following PCR amplification, products were purified using the AMPure XP system. Finally, library quality was assessed on the Agilent Bioanalyzer 2100 system. The resulting libraries were sequenced on the Illumina HiSeq X Ten System in a 2 × 150 bp paired-end mode.
Read alignment and transcript assembly
Figure 1b shows the bioinformatic analysis workflow. The raw sequencing reads10 were subjected to adapter trimming and base quality filtering by fastp v0.7.011. Clean reads obtained were aligned to the human reference genome (GRCh37) using hisat2 v2.1.012 under default settings. Following alignment, the quality of each RNA-seq dataset was assessed through a variety of metrics generated by QoRTs13. Transcript de novo assembly for each sample was performed using StringTie v1.3.4b14 under default settings with the guidance of a reference annotation (GENCODE GRCh37 release 27, -G option). The assembled transcripts of all samples were merged into a single file using the merge function of StringTie with the reference annotation provided (-G option). Other parameters were set to defaults (-m 50 -T 1 -f 0.01 -g 250).
Novel lncRNA gene prediction
The transcripts without matched known transcript information in the StringTie merge output were predicted to be from novel lncRNA genes based on the following criteria: (1) the novel transcripts assembled must have definite strand information; (2) the transcripts must have more than one exon; (3) the transcripts must be more or equal to 200 bp in length; and (4) the coding potential of the transcripts were predicted using CPC215, and only the transcripts labelled as “noncoding” in the output were kept. We ultimately got 205 novel lncRNA genes (ALL_GENE_EXPR_DEG_ANALYSIS.xlsx)16.
Expression abundance quantification
All coding genes and lncRNA genes, including predicted novel lncRNA, lincRNA, sense intronic lncRNA, sense overlapping lncRNA and antisense lncRNA genes, were incorporated in expression abundance quantification (stringtie_merged.strand.lncRNA.proteincoding.gtf)16. Firstly, the transcript sequences (stringtie_merged.strand.lncRNA.proteincoding.fa)16 were extracted from the reference genome using gffread (https://github.com/gpertea/gffread). Then, the expression of the transcripts was quantified with kallisto v0.43.117 under default settings. For comparison among samples, transcript abundance for each sample was normalized with Transcripts Per Million (TPM)18. The expression of each gene was determined by aggregating the expression of all corresponding transcript isoforms. Along with transcript abundance estimates, 100 bootstraps per sample were generated (kallisto quant –b 100), which serve as proxies for technical replicates. Figure 2a,b show the expression profiles of coding genes and lncRNA genes in each sample, respectively. Based on the expression of coding genes, hierarchical clustering analysis revealed distinct expression landscapes between the normal and HCM groups for both coding and lncRNA genes. However, samples from each of the three HCM groups were not clustered together, indicating that there may be no significant difference in transcriptome among HCM patients with different genetic backgrounds at least in the sampling stage.
Expression profiles of coding and lncRNA genes. (a) Hierarchical clustering of the samples from the three HCM groups and the normal group based on the expression of coding genes. (b) Hierarchical clustering of the samples from the three HCM groups and the normal group based on the expression of lncRNA genes. In a and b, each row represents a gene, and each column represents a sample. For better visualization, only the expression of 1,000 randomly selected genes are displayed on the heatmap. (c) Volcano plot showing the differentially expressed coding genes between HCM and normal groups. (d) Volcano plot showing the differentially expressed lncRNA genes between HCM and normal groups. In c and d, dots coloured in light red or light blue denote statistically and biologically significant genes being up-regulated or down-regulated, respectively. The dot size reflects the absolute fold change. Only the top 30 DEGs were labelled with gene symbols.
Differential expression analysis
Following quantification, the identification of differentially expressed genes (DEGs) between HCM and normal samples was performed using sleuth v0.29.019, which could leverage the bootstraps of kallisto to correct for technical variation. The biological significance threshold was set to a fold change of ±2 fold, and the statistical significance threshold was set to a q-value of 0.05 (−log10 q-value > 1.3). Only genes that achieved both biological and statistical significance were considered as DEGs. We identified 132 and 241 coding genes up-regulated and down-regulated in HCM versus normal samples, respectively (Fig. 2c). We also found 67 and 83 lncRNA genes up-regulated and down-regulated in HCM versus normal samples, respectively (Fig. 2d). We made available the useful information for each sample, including the expression abundance of each gene, testing statistics and DEGs (ALL_GENE_EXPR_DEG_ANALYSIS.xlsx)16.
Technical Validation
After quality control, the number of sequenced bases was over 11 Gb in all samples, and the Q20 (the percentage of bases with Phred-scaled quality score ≥20) was over 97% in all samples (Q30 over 93%), indicating that the base quality was sufficiently high for downstream analyses (Table 1). When aligning the clean reads to human reference genome, the overall alignment rate was high (over 97%) in all samples, suggesting little contamination from microorganisms (Table 1).
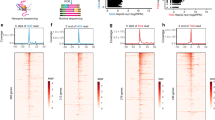
Taking advantage of QoRTs13, a toolkit for quality assessment of RNA-seq dataset, we made cross-comparisons of samples to identify any outliers or systematic errors associated with biological conditions, i.e., different groups (Fig. 3a–f). Figure 3a shows the distribution curve of estimated insert size for each sample. We found that the curves were relatively smooth (no “spikes”) and consistent across samples and conditions, reflecting little technical bias across samples. Figure 3b shows the gene body coverage profile for each sample, and no significant 3’ bias was found, indicating that the datasets were not affected by RNA degradation. Figure 3c shows the read mapping rates for different location categories in each sample, from which we did not observe any outlier within each condition, suggesting consistency across samples in terms of alignment. Similarly, we did not observe a disproportionate identification of novel splice junctions in one sample or condition (Fig. 3d). Except for the nucleotide composition bias in the first few cycles that normally occur in Illumina RNA-seq data, the base composition was quite uniform across all other cycles (Fig. 3e). Figure 3f shows the alignment soft clipping rate by cycle in each sample. We did not observe any “spikes” in the curves for all samples and the clipping profiles were generally consistent across samples and conditions. To visualize the high-dimensional transcriptomic datasets, we performed dimension reduction with principle component analysis (PCA). Consistent with the observation in the hierarchical clustering analysis (Fig. 2a,b), we found that all HCM samples clustered together and were distant from normal samples (Fig. 3g), thus suggesting that our data are suitable for differential expression analysis. As expected, the transcriptomic variance among samples was found to be more significant in the normal condition than the diseased HCM condition.
Quality assessment of the RNA-seq dataset. (a) Distribution curve of estimated insert size for each sample. (b) Gene body coverage profile for each sample. Only the genes in the upper-middle quartile by read-count are considered. (c) Read mapping rates for different location categories in each sample. Unique Gene: exons of only one gene; Unique Gene UTR: UTRs of only one gene; Ambig Gene: exons of more than one gene; No Gene: a region without annotated genes; No Gene, Intronic: a region bridged by an annotated splice junction; No gene, 1 kb from gene: 1 kilobase from the nearest annotated gene; No gene, 10 kb from gene: 10 kilobases from the nearest annotated gene; No gene, middle of nowhere: more than 10 kilobases from the nearest annotated gene (d) Number of splice junctions for different categories of each sample. “1–3 reads” means the junction locus is covered by 1–3 read-pairs. (e) Nucleotide rate by cycle for aligned bases in each sample. Nucleotide types are differentiated by colour. Sample groups are differentiated by shape. (f) Alignment soft clipping rate by cycle in each sample. (a–f) Plots are generated by QoRTs. (g) PCA for visualizing the high-dimensional expression datasets.
Taken together, we presented a high-quality dataset that was suitable for differential expression and splicing analysis of both coding and lncRNA genes in myocardial tissues between HCM and normal conditions.
Code Availability
The code for processing the data from raw sequencing reads to DEGs is available within figshare (CODE_for_RNA-seq.sh)16.
References
Ho, C. Y. Hypertrophic cardiomyopathy in 2012. Circulation 125, 1432–1438 (2012).
Geske, J. B., Ommen, S. R. & Gersh, B. J. Hypertrophic cardiomyopathy: clinical update. JACC Heart Fail 6, 364–375 (2018).
Walsh, R. et al. Defining the genetic architecture of hypertrophic cardiomyopathy: re-evaluating the role of non-sarcomeric genes. Eur. Heart J. 38, 3461–3468 (2017).
Mattick, J. S. & Rinn, J. L. Discovery and annotation of long noncoding RNAs. Nat. Struct. Mol. Biol. 22, 5–7 (2015).
Greco, S., Salgado Somoza, A., Devaux, Y. & Martelli, F. Long Noncoding RNAs and Cardiac Disease. Antioxid. Redox Signal. 29, 880–901 (2018).
Rinn, J. L. & Chang, H. Y. Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 81, 145–166 (2012).
Shen, S., Jiang, H., Bei, Y., Xiao, J. & Li, X. Long non-coding RNAs in cardiac remodeling. Cell. Physiol. Biochem. 41, 1830–1837 (2017).
Yang, W., Li, Y., He, F. & Wu, H. Microarray profiling of long non-coding RNA (lncRNA) associated with hypertrophic cardiomyopathy. BMC Cardiovasc. Disord. 15, 62 (2015).
Mills, J. D., Kawahara, Y. & Janitz, M. Strand-specific RNA-seq provides greater resolution of transcriptome profiling. Curr. Genomics 14, 173–181 (2013).
NCBI Sequence Read Archive, https://identifiers.org/ncbi/insdc.sra:SRP186138 (2019).
Chen, S., Zhou, Y., Chen, Y. & Gu, J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890 (2018).
Kim, D., Langmead, B. & Salzberg, S. L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357 (2015).
Hartley, S. W. & Mullikin, J. C. QoRTs: a comprehensive toolset for quality control and data processing of RNA-Seq experiments. BMC Bioinformatics 16, 224 (2015).
Pertea, M. et al. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33, 290 (2015).
Kang, Y. J. et al. CPC2: a fast and accurate coding potential calculator based on sequence intrinsic features. Nucleic Acids Res. 45, W12–W16 (2017).
Liu, X. Long non-coding RNA and mRNA profiling using RNA sequencing in human hypertrophic cardiomyopathy. figshare, https://doi.org/10.6084/m9.figshare.c.4407284.v1 (2019).
Bray, N. L., Pimentel, H., Melsted, P. & Pachter, L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 34, 525–527 (2016).
Liu, X. & Zhou, Z. Long non-coding and protein-coding RNA profiling using strand-specific RNA-seq in human hypertrophic cardiomyopathy. Gene Expression Omnibus, https://identifiers.org/geo:GSE130036 (2019).
Pimentel, H., Bray, N. L., Puente, S., Melsted, P. & Pachter, L. Differential analysis of RNA-seq incorporating quantification uncertainty. Nat. Methods 14, 687–690 (2017).
Acknowledgements
This work is supported by grants of the National Natural Science Foundation of China (81570276), Beijing Science and Technology Program (Z161100000516154), the CAMS Initiative for Innovative Medicine (2016-I2M-1-016) and the Post-doctoral International Exchange Project (2018-BSH04).
Author information
Authors and Affiliations
Contributions
Y.M. participated in the experimental design. K.Y., T.L., B.H. and Xuewen L. performed RNA isolation and library preparation. W.L., W.C., Y.Z. and C.Z. gave suggestions on data analysis. Xuanyu L. analysed the data and wrote the manuscript. S.W. and Z.Z. conceived the project.
Corresponding authors
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
ISA-Tab metadata file
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
The Creative Commons Public Domain Dedication waiver http://creativecommons.org/publicdomain/zero/1.0/ applies to the metadata files associated with this article.
About this article
Cite this article
Liu, X., Ma, Y., Yin, K. et al. Long non-coding and coding RNA profiling using strand-specific RNA-seq in human hypertrophic cardiomyopathy. Sci Data 6, 90 (2019). https://doi.org/10.1038/s41597-019-0094-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41597-019-0094-6
This article is cited by
-
Lineage-specific regulatory changes in hypertrophic cardiomyopathy unraveled by single-nucleus RNA-seq and spatial transcriptomics
Cell Discovery (2023)
-
Single-cell RNA sequencing identifies an Il1rn+/Trem1+ macrophage subpopulation as a cellular target for mitigating the progression of thoracic aortic aneurysm and dissection
Cell Discovery (2022)
-
Single-nucleus profiling of human dilated and hypertrophic cardiomyopathy
Nature (2022)
-
A critical approach for successful use of circulating microRNAs as biomarkers in cardiovascular diseases: the case of hypertrophic cardiomyopathy
Heart Failure Reviews (2022)
-
Therapeutic benefits of niraparib tosylate as radio sensitizer in esophageal squamous cell carcinoma: an in vivo and in vitro preclinical study
Clinical and Translational Oncology (2022)