Abstract
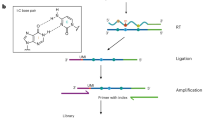
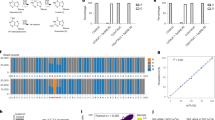
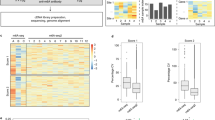
N6-methyladenosine (m6A) is the most abundant mRNA modification in mammalian cells, regulating many physiological processes. Here we describe a method for base-resolution, quantitative m6A sequencing in the whole transcriptome. The enzyme and small-molecule cofactor used in this protocol are prepared by recombinant protein expression and organic synthesis, respectively. Then the library can be prepared from various types of RNA samples using a ligation-based strategy, with m6A modifications being labeled by the enzyme and cofactor. Detailed instructions on ensuing data analysis are also included in this protocol. The method generates highly reproducible results, uncovering 31,233–129,263 sites using as little as 2 ng of poly A+ RNA. These identified sites correspond well with previous m6A profiling results, covering over 65% of peaks detected by the antibody-based approaches. Compared with other currently available methods, this method can be applied to various types of biological samples, including fresh and frozen tissues as well as formalin-fixed paraffin-embedded samples, providing a quantitative method to uncover new insights into m6A biology. The protocol requires basic expertise in molecular biology, recombinant protein expression and organic synthesis. The whole protocol can be done in 15 days, with the library preparation taking 5 days.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Code availability
Code for assessing the quality, reliability and features of the identified m6A sites is available on GitHub (https://github.com/y9c/m6A-SACseq).
Change history
22 June 2023
A Correction to this paper has been published: https://doi.org/10.1038/s41596-023-00862-3
References
Fu, Y. & He, C. Nucleic acid modifications with epigenetic significance. Curr. Opin. Chem. Biol. 16, 516–524 (2012).
Alarcón, C. R., Lee, H., Goodarzi, H., Halberg, N. & Tavazoie, S. F. N6-methyladenosine marks primary microRNAs for processing. Nature 519, 482–485 (2015).
Liu, N. et al. N6-methyladenosine alters RNA structure to regulate binding of a low-complexity protein. Nucleic Acids Res. 45, 6051–6063 (2017).
Wang, X. et al. N6-methyladenosine modulates messenger RNA translation efficiency. Cell 161, 1388–1399 (2015).
Wang, X. et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505, 117–120 (2014).
Zhou, K. I. et al. N6-methyladenosine modification in a long noncoding RNA hairpin predisposes its conformation to protein binding. J. Mol. Biol. 428, 822–833 (2016).
Liu, J. et al. A METTL3–METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 10, 93–95 (2014).
Ping, X.-L. et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 24, 177–189 (2014).
Śledź, P. & Jinek, M. Structural insights into the molecular mechanism of the m6A writer complex. eLife 5, e18434. (2016).
Wang, X. et al. Structural basis of N6-adenosine methylation by the METTL3–METTL14 complex. Nature 534, 575–578 (2016).
Wang, Y. et al. N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat. Cell Biol. 16, 191–198 (2014).
Wei, C.-M., Gershowitz, A. & Moss, B. Methylated nucleotides block 5′ terminus of HeLa cell messenger RNA. Cell 4, 379–386 (1975).
Fu, Y., Dominissini, D., Rechavi, G. & He, C. Gene expression regulation mediated through reversible m6A RNA methylation. Nat. Rev. Genet. 15, 293 (2014).
He, P. C. & He, C. m6A RNA methylation: from mechanisms to therapeutic potential. EMBO J. 40, (2021).
Yang, Y., Hsu, P. J., Chen, Y.-S. & Yang, Y.-G. Dynamic transcriptomic m6A decoration: writers, erasers, readers and functions in RNA metabolism. Cell Res. 28, 616–624 (2018).
Dominissini, D. et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485, 201–206 (2012).
Meyer, K. D. et al. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell 149, 1635–1646 (2012).
Linder, B. et al. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat. Methods 12, 767–772 (2015).
Meyer, K. D. DART-seq: an antibody-free method for global m6A detection. Nat. Methods 16, 1275–1280 (2019).
Zhang, Z. et al. Single-base mapping of m6A by an antibody-independent method. Sci. Adv. 5, eaax0250 (2019).
Garcia-Campos, M. A. et al. Deciphering the “m6A code” via antibody-independent quantitative profiling. Cell 178, 731–747.e16 (2019).
Shu, X. et al. A metabolic labeling method detects m6A transcriptome-wide at single base resolution. Nat. Chem. Biol. 16, 887–895 (2020).
Hu, L. et al. m6A RNA modifications are measured at single-base resolution across the mammalian transcriptome. Nat. Biotechnol. 40, 1210–1219 (2022).
O’Farrell, H. C., Musayev, F. N., Scarsdale, J. N. & Rife, J. P. Binding of adenosine-based ligands to the MjDim1 rRNA methyltransferase: implications for reaction mechanism and drug design. Biochemistry 49, 2697–2704 (2010).
O’Farrell, H. C., Pulicherla, N., Desai, P. M. & Rife, J. P. Recognition of a complex substrate by the KsgA/Dim1 family of enzymes has been conserved throughout evolution. RNA 12, 725–733 (2006).
Shu, X. et al. N6-allyladenosine: a new small molecule for RNA labeling identified by mutation assay. J. Am. Chem. Soc. 139, 17213–17216 (2017).
Golinelli, M.-P. & Hughes, S. H. Nontemplated nucleotide addition by HIV-1 reverse transcriptase. Biochemistry 41, 5894–5906 (2002).
Davis, W. R., Gabbara, S., Hupe, D. & Peliska, J. A. Actinomycin D inhibition of DNA strand transfer reactions catalyzed by HIV-1 reverse transcriptase and nucleocapsid protein. Biochemistry 37, 14213–14221 (1998).
Song, Y., Liu, K. J. & Wang, T.-H. Elimination of ligation dependent artifacts in T4 RNA ligase to achieve high efficiency and low bias microRNA capture. PLoS ONE 9, e94619 (2014).
Rusche, J. R. & Howard-Flanders, P. Hexamine cobalt chloride promotes intermolecular ligation of blunt end DNA fragments by T4 DNA ligase. Nucleic Acids Res. 13, 1997–2008 (1985).
Zhuang, F., Fuchs, R. T., Sun, Z., Zheng, Y. & Robb, G. B. Structural bias in T4 RNA ligase-mediated 3′-adapter ligation. Nucleic Acids Res. 40, e54–e54 (2012).
Wang, K., Peng, J. & Yi, C. The m6A consensus motif provides a paradigm of epitranscriptomic studies. Biochemistry 60, 3410–3412 (2021).
Wei, C.-M. & Moss, B. Nucleotide sequences at the N6-methyladenosine sites of HeLa cell messenger ribonucleic acid. Biochemistry 16, 1672–1676 (1977).
Ke, S. et al. A majority of m6A residues are in the last exons, allowing the potential for 3′ UTR regulation. Genes Dev. 29, 2037–2053 (2015).
Wang, Y., Xiao, Y., Dong, S., Yu, Q. & Jia, G. Antibody-free enzyme-assisted chemical approach for detection of N6-methyladenosine. Nat. Chem. Biol. 16, 896–903 (2020).
Zhang, Z. et al. Systematic calibration of epitranscriptomic maps using a synthetic modification-free RNA library. Nat. Methods 18, 1213–1222 (2021).
Zhang, L.-S. et al. Transcriptome-wide mapping of internal N7-methylguanosine methylome in mammalian mRNA. Mol. Cell 74, 1304–1316.e8 (2019).
Ke, S. et al. m 6 A mRNA modifications are deposited in nascent pre-mRNA and are not required for splicing but do specify cytoplasmic turnover. Genes Dev. 31, 990–1006 (2017).
Maden, B. E. H. Locations of methyl groups in 28S rRNA of Xenopus laevis and man: clustering in the conserved core of molecule. J. Mol. Biol. 201, 289–314 (1988).
Maden, B. E. H. Identification of the locations of the methyl groups in 18S ribosomal RNA from Xenopus laevis and man. J. Mol. Biol. 189, 681–699 (1986).
Yang, X. et al. N6-methyladenine modification in noncoding RNAs and its function in cancer. Biomarker Res. 8, (2020).
Mossessova, E. & Lima, C. D. Ulp1-SUMO crystal structure and genetic analysis reveal conserved interactions and a regulatory element essential for cell growth in yeast. Mol. Cell 5, 865–876 (2000).
Liu, J. et al. N6-methyladenosine of chromosome-associated regulatory RNA regulates chromatin state and transcription. Science 367, 580–586 (2020).
Tegowski, M., Flamand, M. N. & Meyer, K. D. scDART-seq reveals distinct m6A signatures and mRNA methylation heterogeneity in single cells. Mol. Cell 82, 868–878.e10 (2022).
Acknowledgements
The authors thank Dr. P. Faber and the University of Chicago Genomics Facility for sequencing support. The authors also thank L. S. Zhang and T. Pan’s lab for discussions. This work was supported by NIH RM1 HG008935, R01CA251150 and the Howard Hughes Medical Institute (C.H. is an investigator of the Howard Hughes Medical Institute).
Author information
Authors and Affiliations
Contributions
R.G. and C.Y. contributed equally to this work. R.G. developed experimental procedures and performed the experiments. C.Y. developed the analysis pipeline and performed data analysis. Y.P. and S.L. helped in the development of the analysis pipeline and provided valuable discussions on the development of the method. Q.D. synthesized the probes. R.G., Y.Z., Q.D. and P.W. all participated in the synthesis and purification of allyl-SAM. L.H. provided valuable discussions on drafting the manuscript. R.G., C.Y. and C.H. wrote and edited the manuscript.
Corresponding authors
Ethics declarations
Competing interests
A patent application for m6A-SAC-seq has been filed by the University of Chicago. C.H. is a scientific founder and a scientific advisory board member of Accent Therapeutics, Inc. and Inferna Green, Inc.
Peer review
Peer review information
Nature Protocols thanks the anonymous reviewers for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key reference using this protocol
Hu, L. et al. Nat. Biotechnol. 40, 1210–1219 (2022): https://doi.org/10.1038/s41587-022-01243-z
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ge, R., Ye, C., Peng, Y. et al. m6A-SAC-seq for quantitative whole transcriptome m6A profiling. Nat Protoc 18, 626–657 (2023). https://doi.org/10.1038/s41596-022-00765-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-022-00765-9
This article is cited by
-
Decoding epitranscriptomic regulation of viral infection: mapping of RNA N6-methyladenosine by advanced sequencing technologies
Cellular & Molecular Biology Letters (2024)
-
Profiling of RNA-binding protein binding sites by in situ reverse transcription-based sequencing
Nature Methods (2024)
-
MePMe-seq: antibody-free simultaneous m6A and m5C mapping in mRNA by metabolic propargyl labeling and sequencing
Nature Communications (2023)
-
RNA modifications in hematological malignancies
International Journal of Hematology (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.