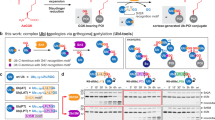
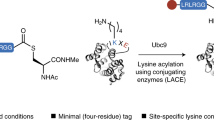
Abstract
Ubiquitination regulates almost every life process of eukaryotes. The study of the ubiquitin (Ub) coupling or decoupling process and the interaction study of Ub–Ub binding protein have always been the central focus. However, such studies are challenging, owing to the transient nature of Ub-coupling enzymes and deubiquitinases in the reactions, as well as the difficulty in preparing large quantities of polyubiquitinated samples. Here we describe a recently developed strategy for the efficient preparation of analogs of Ub chains and analogs for Ub coupling and uncoupling intermediates, which facilitate the study of the ubiquitination process. The strategy includes mainly the following steps: (i) the bifunctional molecule conjugation on the only cysteine (Cys) residue of a target protein (usually a Ub or Ub-conjugating enzyme), exposing an orthogonal reactive site for native chemical ligation; (ii) covalent ligation with a Ub-derived thioester, exposing a free sulfhydryl; and (iii) (optional) a disulfide bond formation with a substrate protein (mainly with only one Cys protein) through nonactivity-based cross-linking or with a deubiquitinase (mainly with several Cys residues) through activity-based cross-linking. When the bifunctional molecule and target proteins are obtained, the final products can be prepared in milligram quantities within 2 weeks.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
The data associated with these results are included in the Supplementary Information and Source Data files.
References
Yau, R. & Rape, M. The increasing complexity of the ubiquitin code. Nat. Cell. Biol. 18, 579–586 (2016).
Oh, E., Akopian, D. & Rape, M. Principles of ubiquitin-dependent signaling. Annu. Rev. Cell. Dev. Biol. 34, 137–162 (2018).
Schulman, B. A. & Harper, J. W. Ubiquitin-like protein activation by E1 enzymes: the apex for downstream signalling pathways. Nat. Rev. Mol. Cell. Biol. 10, 319–331 (2009).
Ye, Y. & Rape, M. Building ubiquitin chains: E2 enzymes at work. Nat. Rev. Mol. Cell. Biol. 10, 755–764 (2009).
Zheng, N. & Shabek, N. Ubiquitin ligases: structure, function, and regulation. Annu. Rev. Biochem. 86, 129–157 (2017).
Clague, M. J., Urbe, S. & Komander, D. Breaking the chains: deubiquitylating enzyme specificity begets function. Nat. Rev. Mol. Cell Biol. 20, 338–352 (2019).
Lange, S. M., Armstrong, L. A. & Kulathu, Y. Deubiquitinases: from mechanisms to their inhibition by small molecules. Mol. Cell 82, 15–29 (2022).
Komander, D. & Rape, M. The ubiquitin code. Annu. Rev. Biochem. 81, 203–229 (2012).
Husnjak, K. & Dikic, I. Ubiquitin-binding proteins: decoders of ubiquitin-mediated cellular functions. Annu. Rev. Biochem. 81, 291–322 (2012).
Senft, D., Qi, J. & Ronai, Z. A. Ubiquitin ligases in oncogenic transformation and cancer therapy. Nat. Rev. Cancer 18, 69–88 (2018).
Schmidt, M. F., Gan, Z. Y., Komander, D. & Dewson, G. Ubiquitin signalling in neurodegeneration: mechanisms and therapeutic opportunities. Cell Death Differ. 28, 570–590 (2021).
Cruz Walma, D. A., Chen, Z., Bullock, A. N. & Yamada, K. M. Ubiquitin ligases: guardians of mammalian development. Nat. Rev. Mol. Cell Biol. 23, 350–367 (2022).
Wertz, I. E. & Wang, X. From discovery to bedside: targeting the ubiquitin system. Cell Chem. Biol. 26, 156–177 (2019).
Buetow, L. & Huang, D. T. Structural insights into the catalysis and regulation of E3 ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 17, 626–642 (2016).
Sui, X. et al. Development and application of ubiquitin-based chemical probes. Chem. Sci. 11, 12633–12646 (2020).
Henneberg, L. T. & Schulman, B. A. Decoding the messaging of the ubiquitin system using chemical and protein probes. Cell Chem. Biol. 28, 889–902 (2021).
Zhang, X. et al. An interaction landscape of ubiquitin signaling. Mol. Cell 65, 941–955 (2017).
Zhao, X. et al. Identification of proteins interacting with ubiquitin chains. Angew. Chem. Int. Ed. Engl. 56, 15764–15768 (2017).
Zhang, X. et al. Proteome-wide identification of ubiquitin interactions using UbIA-MS. Nat. Protoc. 13, 530–550 (2018).
Lutz, J., Hollmuller, E., Scheffner, M., Marx, A. & Stengel, F. The length of a ubiquitin chain: a general factor for selective recognition by ubiquitin-binding proteins. Angew. Chem. Int. Ed. Engl. 59, 12371––12375 (2020).
Streich, F. C. Jr. & Lima, C. D. Capturing a substrate in an activated RING E3/E2–SUMO complex. Nature 536, 304–308 (2016).
Baek, K. et al. NEDD8 nucleates a multivalent cullin–RING–UBE2D ubiquitin ligation assembly. Nature 578, 461–466 (2020).
Horn-Ghetko, D. et al. Ubiquitin ligation to F-box protein targets by SCF–RBR E3–E3 super-assembly. Nature 590, 671–676 (2021).
Pao, K. C. et al. Activity-based E3 ligase profiling uncovers an E3 ligase with esterification activity. Nature 556, 381–385 (2018).
Mabbitt, P. D. et al. Structural basis for RING-Cys-Relay E3 ligase activity and its role in axon integrity. Nat. Chem. Biol. 16, 1227–1236 (2020).
Liang, L. J. et al. Chemical synthesis of activity-based E2-ubiquitin probes for the structural analysis of E3 ligase-catalyzed transthiolation. Angew. Chem. Int. Ed. Engl. 60, 17171–17177 (2021).
Pan, M. et al. Chemical synthesis of structurally defined phosphorylated ubiquitins suggests impaired parkin activation by phosphorylated ubiquitins with a non-phosphorylated distal unit. CCS Chem. 1, 476–489 (2019).
Mevissen, T. E. T. et al. Molecular basis of Lys11-polyubiquitin specificity in the deubiquitinase Cezanne. Nature 538, 402–405 (2016).
Zheng, Q. et al. An E1-catalyzed chemoenzymatic strategy to isopeptide-N-ethylated deubiquitylase-resistant ubiquitin probes. Angew. Chem. Int. Ed. Engl. 59, 13496–13501 (2020).
Pan, M. et al. Structural insights into Ubr1-mediated N-degron polyubiquitination. Nature 600, 334–338 (2021).
Pan, M. et al. Chemical protein synthesis enabled mechanistic studies on the molecular recognition of K27-linked ubiquitin chains. Angew. Chem. Int. Ed. Engl. 58, 2627–2631 (2019).
Varshavsky, A. N-degron and C-degron pathways of protein degradation. Proc. Natl Acad. Sci. USA 116, 358–366 (2019).
Sherpa, D., Chrustowicz, J. & Schulman, B. A. How the ends signal the end: regulation by E3 ubiquitin ligases recognizing protein termini. Mol. Cell 82, 1424–1438 (2022).
Mevissen, T. E. et al. OTU deubiquitinases reveal mechanisms of linkage specificity and enable ubiquitin chain restriction analysis. Cell 154, 169–184 (2013).
Maity, S. K., Jbara, M., Laps, S. & Brik, A. Efficient palladium-assisted one-pot deprotection of (acetamidomethyl)cysteine following native chemical ligation and/or desulfurization to expedite chemical protein synthesis. Angew. Chem. Int. Ed. Engl. 55, 8108–8112 (2016).
Pan, M. et al. Quasi-racemic X-ray structures of K27-linked ubiquitin chains prepared by total chemical synthesis. J. Am. Chem. Soc. 138, 7429–7435 (2016).
van Tilburg, G. B. A. et al. K27-linked diubiquitin inhibits UCHL3 via an unusual kinetic trap. Cell Chem. Biol. 28, 191–201 (2021).
Flierman, D. et al. Non-hydrolyzable diubiquitin probes reveal linkage-specific reactivity of deubiquitylating enzymes mediated by S2 pockets. Cell Chem. Biol. 23, 472–482 (2016).
Ye, Y. et al. Ubiquitin chain conformation regulates recognition and activity of interacting proteins. Nature 492, 266–270 (2012).
Chu, G. C. et al. Cysteine-aminoethylation-assisted chemical ubiquitination of recombinant histones. J. Am. Chem. Soc. 141, 3654–3663 (2019).
Brown, N. G. et al. Dual RING E3 architectures regulate multiubiquitination and ubiquitin chain elongation by APC/C. Cell 165, 1440–1453 (2016).
de la Torre, D. & Chin, J. W. Reprogramming the genetic code. Nat. Rev. Genet. 22, 169–184 (2021).
Tang, S. et al. Practical chemical synthesis of atypical ubiquitin chains by using an isopeptide-linked Ub isomer. Angew. Chem. Int. Ed. Engl. 56, 13333–13337 (2017).
Fairhead, M. & Howarth, M. Site-specific biotinylation of purified proteins using BirA. Methods Mol. Biol. 1266, 171–184 (2015).
Adams, A. L. et al. Cysteine promoted C-terminal hydrazinolysis of native peptides and proteins. Angew. Chem. Int. Ed. Engl. 52, 13062–13066 (2013).
Wang, X. A., Kurra, Y., Huang, Y., Lee, Y. J. & Liu, W. R. E1-catalyzed ubiquitin C-terminal amidation for the facile synthesis of deubiquitinase substrates. ChemBioChem 15, 37–41 (2014).
Qu, Q. et al. A highly efficient synthesis of polyubiquitin chains. Adv. Sci. 5, 1800234 (2018).
Bachmair, A., Finley, D. & Varshavsky, A. In vivo half-life of a protein is a function of its amino-terminal residue. Science 234, 179–186 (1986).
Zheng, J. S., Tang, S., Qi, Y. K., Wang, Z. P. & Liu, L. Chemical synthesis of proteins using peptide hydrazides as thioester surrogates. Nat. Protoc. 8, 2483–2495 (2013).
Tang, S. et al. Chemical synthesis of membrane proteins by the removable backbone modification method. Nat. Protoc. 12, 2554–2569 (2017).
Acknowledgements
We thank H. Deng, X. Meng and X. Tian in the Protein Chemistry and Proteomics Facility, Tsinghua University Technology Center for Protein Research, for protein MS analysis. This work was supported by Shanghai Rising-Star Program (no. 22QA1404900), the National Key R&D Program of China (no. 2021YFC2100201) and the National Natural Science Foundation of China (nos. 22277073, 22207070, 91849129 and 22077078). Q.Z. thanks the funding by the National Facility for Translational Medicine (Shanghai).
Author information
Authors and Affiliations
Contributions
M.P. and H.H. led the project and supervised the project. Q.Z. and M.P. designed all experiments and prepared the manuscript. G.C., Y.Y. and H.H. synthesized the bifunctional molecule. T.W., J.M. and C.Z. synthesized the free Ub chain mimics. J.M. and L.L. synthesized the Ub coupling intermediate mimics. T.W. and Y.J. synthesized the Ub decoupling intermediate mimics. Q.Z. and T.W. performed the experiments on HDX-MS and cross-linking site identification. All authors contributed to the writing of the manuscript and approved the final version.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Protocols thanks the anonymous reviewers for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key references using this protocol
Pan, M. et. al. Nature 600, 334–338 (2021): https://doi.org/10.1038/s41586-021-04097-8.
Zheng, Q. et al. Angew. Chem. In. Ed. 59, 13496–13501 (2020): https://doi.org/10.1002/anie.202002974.
Extended data
Extended Data Fig. 1 Intermediate products of the Acm-deprotection reaction and CAET-assisted NCL reaction for preparation of Ubc2-Ub-K48Ub-Y/degron.
a, RP-HPLC trace (214 nm) and ESI-MS of purified product 6. b, RP-HPLC trace (214 nm) and ESI-MS of purified product 8.
Extended Data Fig. 2 LC–MS characterization of Ub-K27C-CAET module and monitoring the reaction efficiency of the DUB intermediate mimic.
LC–MS characterization of Ub-K27C-CAET module and monitoring the reaction efficiency of the DUB intermediate mimic. a, RP-HPLC trace (214 nm) and ESI-MS of purified product 12. b, SDS–PAGE analysis of the activity-based cross-linking between the enzyme Otud2 and K27-CAET-AT2 diUb.
Supplementary information
Supplementary Information
Supplementary Figs. 1–5 and Methods 1 and 2.
Source data
Source Data Fig. 3
Unprocessed gels for Fig. 3.
Source Data Fig. 4
Unprocessed gels for Fig. 4c.
Source Data Fig. 5
Unprocessed gels for Fig. 5e.
Source Data Extended Data Fig. 2
Unprocessed gels for Extended Data Fig. 2b.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zheng, Q., Wang, T., Mao, J. et al. A bifunctional molecule-assisted synthesis of mimics for use in probing the ubiquitination system. Nat Protoc 18, 530–554 (2023). https://doi.org/10.1038/s41596-022-00761-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-022-00761-z
This article is cited by
-
Structure-guided engineering enables E3 ligase-free and versatile protein ubiquitination via UBE2E1
Nature Communications (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.