Abstract
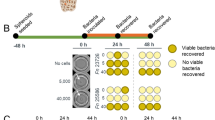
The prevalence of tumor-colonizing bacteria along with advances in synthetic biology are leading to a new generation of living microbial cancer therapies. Because many bacterial systems can be engineered to recombinantly produce therapeutics within tumors, simple and high-throughput experimental platforms are needed to screen the large collections of bacteria candidates and characterize their interactions with cancer cells. Here, we describe a protocol to selectively grow bacteria within the core of tumor spheroids, allowing for their continuous and parallel profiling in physiologically relevant conditions. Specifically, tumor spheroids are incubated with bacteria in a 96-well low-adhesion plate followed by a series of washing steps and an antibiotic selection protocol to confine bacterial growth within the hypoxic and necrotic core of tumor spheroids. This bacteria spheroid coculture (BSCC) system is stable for over 2 weeks, does not require specialized equipment and is compatible with time-lapse microscopy, commercial staining assays and histology that uniquely enable analysis of growth kinetics, viability and spatial distribution of both cellular populations, respectively. We show that the procedure is applicable to multiple tumor cell types and bacterial species by varying protocol parameters and is validated by using animal models. The BSCC platform will allow the study of bacteria–tumor interactions in a continuous manner and facilitate the rapid development of engineered microbial therapies.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
The main data discussed in this protocol were generated as part of the studies published in the supporting primary research papers20,21. Source data are provided for Figs. 4–7. Source data are provided with this paper.
Code availability
The computational code used for image analysis is provided in Supplementary Information.
References
Sepich-Poore, G. D. et al. The microbiome and human cancer. Science 371, eabc4552 (2021).
Nejman, D. et al. The human tumor microbiome is composed of tumor type-specific intracellular bacteria. Science 368, 973–980 (2020).
Zhou, S., Gravekamp, C., Bermudes, D. & Liu, K. Tumour-targeting bacteria engineered to fight cancer. Nat. Rev. Cancer 18, 727–743 (2018).
Harimoto, T. & Danino, T. Engineering bacteria for cancer therapy. Emerg. Top. Life Sci. 3, 623–629 (2019).
Chien, T., Doshi, A. & Danino, T. Advances in bacterial cancer therapies using synthetic biology. Curr. Opin. Syst. Biol. 5, 1–8 (2017).
Bullman, S. et al. Analysis of Fusobacterium persistence and antibiotic response in colorectal cancer. Science 358, 1443–1448 (2017).
Uemura, N. et al. Helicobacter pylori infection and the development of gastric cancer. N. Engl. J. Med. 345, 784–789 (2001).
Kalaora, S. et al. Identification of bacteria-derived HLA-bound peptides in melanoma. Nature 592, 138–143 (2021).
Geller, L. T. et al. Potential role of intratumor bacteria in mediating tumor resistance to the chemotherapeutic drug gemcitabine. Science 357, 1156–1160 (2017).
Pleguezuelos-Manzano, C. et al. Mutational signature in colorectal cancer caused by genotoxic pks+ E. coli. Nature 580, 269–273 (2020).
Lehouritis, P., Springer, C. & Tangney, M. Bacterial-directed enzyme prodrug therapy. J. Control. Release 170, 120–131 (2013).
Isabella, V. M. et al. Development of a synthetic live bacterial therapeutic for the human metabolic disease phenylketonuria. Nat. Biotechnol. 36, 857–864 (2018).
Flickinger, J. C. Jr., Rodeck, U. & Snook, A. E. Listeria monocytogenes as a vector for cancer immunotherapy: current understanding and progress. Vaccines (Basel) 6, 48 (2018).
Toso, J. F. et al. Phase I study of the intravenous administration of attenuated Salmonella typhimurium to patients with metastatic melanoma. J. Clin. Oncol. 20, 142–152 (2002).
Roberts, N. J. et al. Intratumoral injection of Clostridium novyi-NT spores induces antitumor responses. Sci. Transl. Med. 6, 249ra111 (2014).
Nemunaitis, J. et al. Pilot trial of genetically modified, attenuated Salmonella expressing the E. coli cytosine deaminase gene in refractory cancer patients. Cancer Gene Ther. 10, 737–744 (2003).
Helmink, B. A., Khan, M. A. W., Hermann, A., Gopalakrishnan, V. & Wargo, J. A. The microbiome, cancer, and cancer therapy. Nat. Med. 25, 377–388 (2019).
Riglar, D. T. & Silver, P. A. Engineering bacteria for diagnostic and therapeutic applications. Nat. Rev. Microbiol. 16, 214–225 (2018).
Forbes, N. S. Engineering the perfect (bacterial) cancer therapy. Nat. Rev. Cancer 10, 785–794 (2010).
Harimoto, T. et al. Rapid screening of engineered microbial therapies in a 3D multicellular model. Proc. Natl Acad. Sci. USA 116, 9002–9007 (2019).
Chien, T. et al. Enhancing the tropism of bacteria via genetically programmed biosensors. Nat. Biomed. Eng. 6, 94–104 (2022).
Zuniga, A. et al. Engineered L-lactate responding promoter system operating in glucose-rich and anoxic environments. ACS Synth. Biol. 10, 3527–3536 (2021).
Kasper, S. H. et al. Colorectal cancer-associated anaerobic bacteria proliferate in tumor spheroids and alter the microenvironment. Sci. Rep. 10, 5321 (2020).
Yano, S. et al. Tumor-targeting Salmonella typhimurium A1-R decoys quiescent cancer cells to cycle as visualized by FUCCI imaging and become sensitive to chemotherapy. Cell Cycle 13, 3958–3963 (2014).
Gao, S. et al. Development of oxytolerant Salmonella typhimurium using radiation mutation technology (RMT) for cancer therapy. Sci. Rep. 10, 3764 (2020).
Sadaghian Sadabad, M. et al. A simple coculture system shows mutualism between anaerobic faecalibacteria and epithelial Caco-2 cells. Sci. Rep. 5, 17906 (2015).
Zheng, D. W. et al. Optically-controlled bacterial metabolite for cancer therapy. Nat. Commun. 9, 1680 (2018).
Wang, S. B. et al. Bacteria-assisted selective photothermal therapy for precise tumor inhibition. Adv. Funct. Mater. 29, 1904093 (2019).
Wang, X. N., Niu, M. T., Fan, J. X., Chen, Q. W. & Zhang, X. Z. Photoelectric bacteria enhance the in situ production of tetrodotoxin for antitumor therapy. Nano Lett. 21, 4270–4279 (2021).
Song, J. et al. A microfluidic device for studying chemotaxis mechanism of bacterial cancer targeting. Sci. Rep. 8, 6394 (2018).
Hong, J. W., Song, S. & Shin, J. H. A novel microfluidic co-culture system for investigation of bacterial cancer targeting. Lab Chip 13, 3033–3040 (2013).
Din, M. O. et al. Synchronized cycles of bacterial lysis for in vivo delivery. Nature 536, 81–85 (2016).
Zhang, W., Mao, S., He, Z., Wu, Z. & Lin, J. M. In situ monitoring of fluid shear stress enhanced adherence of bacteria to cancer cells on microfluidic chip. Anal. Chem. 91, 5973–5979 (2019).
Mokrani, N. et al. Magnetotactic bacteria penetration into multicellular tumor spheroids for targeted therapy. Annu. Int. Conf. IEEE Eng. Med. Biol. Soc. 2010, 4371–4374 (2010).
Suh, S. et al. Nanoscale bacteria-enabled autonomous drug delivery system (NanoBEADS) enhances intratumoral transport of nanomedicine. Adv. Sci. (Weinh.) 6, 1801309 (2019).
Kasinskas, R. W. & Forbes, N. S. Salmonella typhimurium specifically chemotax and proliferate in heterogeneous tumor tissue in vitro. Biotechnol. Bioeng. 94, 710–721 (2006).
Deng, Y., Liu, S. Y., Chua, S. L. & Khoo, B. L. The effects of biofilms on tumor progression in a 3D cancer-biofilm microfluidic model. Biosens. Bioelectron. 180, 113113 (2021).
Brackett, E. L., Swofford, C. A. & Forbes, N. S. Microfluidic device to quantify the behavior of therapeutic bacteria in three-dimensional tumor tissue. Methods Mol. Biol. 1409, 35–48 (2016).
Goers, L., Freemont, P. & Polizzi, K. M. Co-culture systems and technologies: taking synthetic biology to the next level. J. R. Soc. Interface 11, 20140065 (2014).
Swofford, C. A., St Jean, A. T., Panteli, J. T., Brentzel, Z. J. & Forbes, N. S. Identification of Staphylococcus aureus α-hemolysin as a protein drug that is secreted by anticancer bacteria and rapidly kills cancer cells. Biotechnol. Bioeng. 111, 1233–1245 (2014).
Quail, D. F. & Joyce, J. A. Microenvironmental regulation of tumor progression and metastasis. Nat. Med. 19, 1423–1437 (2013).
Yu, B. et al. Explicit hypoxia targeting with tumor suppression by creating an “obligate” anaerobic Salmonella Typhimurium strain. Sci. Rep. 2, 436 (2012).
Ryan, R. M. et al. Bacterial delivery of a novel cytolysin to hypoxic areas of solid tumors. Gene Ther. 16, 329–339 (2009).
Friedrich, J., Seidel, C., Ebner, R. & Kunz-Schughart, L. A. Spheroid-based drug screen: considerations and practical approach. Nat. Protoc. 4, 309–324 (2009).
Fong, E. L., Harrington, D. A., Farach-Carson, M. C. & Yu, H. Heralding a new paradigm in 3D tumor modeling. Biomaterials 108, 197–213 (2016).
Osswald, A. et al. Three-dimensional tumor spheroids for in vitro analysis of bacteria as gene delivery vectors in tumor therapy. Microb. Cell Fact. 14, 199 (2015).
Bhave, M. S., Hassanbhai, A. M., Anand, P., Luo, K. Q. & Teoh, S. H. Effect of heat-inactivated Clostridium sporogenes and its conditioned media on 3-dimensional colorectal cancer cell models. Sci. Rep. 5, 15681 (2015).
Co, J. Y., Margalef-Catala, M., Monack, D. M. & Amieva, M. R. Controlling the polarity of human gastrointestinal organoids to investigate epithelial biology and infectious diseases. Nat. Protoc. 16, 5171–5192 (2021).
Zhang, J. et al. Coculture of primary human colon monolayer with human gut bacteria. Nat. Protoc. 16, 3874–3900 (2021).
Aguilar, C. et al. Organoids as host models for infection biology—a review of methods. Exp. Mol. Med. 53, 1471–1482 (2021).
Danino, T., Lo, J., Prindle, A., Hasty, J. & Bhatia, S. N. In vivo gene expression dynamics of tumor-targeted bacteria. ACS Synth. Biol. 1, 465–470 (2012).
Canale, F. P. et al. Metabolic modulation of tumours with engineered bacteria for immunotherapy. Nature 598, 662–666 (2021).
Doi, Y., Wachino, J. I. & Arakawa, Y. Aminoglycoside resistance: the emergence of acquired 16S ribosomal RNA methyltransferases. Infect. Dis. Clin. North Am. 30, 523–537 (2016).
Codini, M. et al. Gentamicin arrests cancer cell growth: the intriguing involvement of nuclear sphingomyelin metabolism. Int. J. Mol. Sci. 16, 2307–2319 (2015).
Lee, C. H., Wu, C. L. & Shiau, A. L. Endostatin gene therapy delivered by Salmonella choleraesuis in murine tumor models. J. Gene Med. 6, 1382–1393 (2004).
Yu, B. et al. Obligate anaerobic Salmonella typhimurium strain YB1 treatment on xenograft tumor in immunocompetent mouse model. Oncol. Lett. 10, 1069–1074 (2015).
Danino, T. et al. Programmable probiotics for detection of cancer in urine. Sci. Transl. Med. 7, 289ra284 (2015).
Acknowledgements
We thank the Herbert Irving Comprehensive Cancer Center Molecular Pathology Shared Resources Facility for help with histological sample processing. We thank W. Mather for assistance with the image analysis. We thank members of the Danino laboratory for reviewing the manuscript. This work was supported by the DoD LC160314 (T.D.), DoD BC160541 (T.D.), NIH 1R01EB029750 (T.D.), NIH F99CA253756 (T.H.) and Honjo International Foundation Scholarship (T.H.).
Author information
Authors and Affiliations
Contributions
T.H. and T.D. conceived and designed the study. T.H., D.D. and T.D. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Protocols thanks Jun Sun and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key references using this protocol
Harimoto, T. et al. Proc. Natl. Acad. Sci. USA 116, 9002–9007 (2019): https://doi.org/10.1073/pnas.1820824116
Chien, T. et al. Nat. Biomed. Eng. 6, 94–104 (2022): https://doi.org/10.1038/s41551-021-00772-3
Supplementary information
Supplementary Information
Supplementary Figs. 1 and 2 and computational code for image analysis
Source data
Source Data Fig. 4
Statistical source data
Source Data Fig. 5
Statistical source data
Source Data Fig. 6
Statistical source data
Source Data Fig. 7
Statistical source data
Rights and permissions
About this article
Cite this article
Harimoto, T., Deb, D. & Danino, T. A rapid screening platform to coculture bacteria within tumor spheroids. Nat Protoc 17, 2216–2239 (2022). https://doi.org/10.1038/s41596-022-00723-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-022-00723-5
This article is cited by
-
Design of combination therapy for engineered bacterial therapeutics in non-small cell lung cancer
Scientific Reports (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.