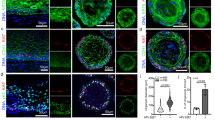
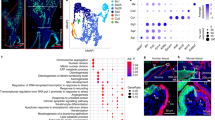
Abstract
The cervix is the gateway to the upper female reproductive tract, connecting the uterus and vagina. It plays crucial roles in fertility and pregnancy maintenance from onset until delivery of the fetus, and prevents pathogen ascension. Compromised functionality of the cervix can lead to disorders, including infertility, chronic infections and cancers. The cervix comprises two regions: columnar epithelium-lined endocervix and stratified squamous epithelium-lined ectocervix, meeting at the squamocolumnar transition zone. So far, two-dimensional cultures of genetically unstable immortalized or cancer cell lines have been primarily used to study cervix biology in vitro. The lack of an in vitro system that reflects the cellular, physiological and functional properties of the two epithelial types has hampered the study of normal physiology, disease development and infection processes. Here we describe a protocol for cell isolation, establishment, long-term culture and expansion of adult epithelial stem cell-derived endocervical and ectocervical organoids from human biopsies and mouse tissue. These two organoid types require unique combinations of growth factors reminiscent of their in vivo tissue niches and different culturing procedures. They recapitulate native three-dimensional tissue architecture and patterning. The protocol to generate these organoids takes 4–6 weeks. We also describe procedures to introduce human papillomavirus oncogenes into the cervical stem cells by genetic manipulation to model cervical cancer and infection of the organoids with the highly prevalent sexually transmitted bacterial pathogen Chlamydia trachomatis. These organoid systems open new possibilities to study cervix biology, infections and cancer evolution, and have potential applications in personalized medicine, drug screening, genome editing and disease modeling.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The main data discussed in this protocol were generated as part of the studies published in the supporting primary research papers by Chumduri et al.4 and Koster et al.13. Source data are provided with this paper.
References
Chumduri, C. & Turco, M. Y. Organoids of the female reproductive tract. J. Mol. Med. 99, 531–553 (2021).
Wira, C. R., Rodriguez-Garcia, M. & Patel, M. V. The role of sex hormones in immune protection of the female reproductive tract. Nat. Rev. Immunol. 15, 217–230 (2015).
Wira, C. R., Grant-Tschudy, K. S. & Crane-Godreau, M. A. Epithelial cells in the female reproductive tract: a central role as sentinels of immune protection. Am. J. Reprod. Immunol. 53, 65–76 (2005).
Chumduri, C. et al. Opposing Wnt signals regulate cervical squamocolumnar homeostasis and emergence of metaplasia. Nat. Cell Biol. 23, 184–197 (2021).
Martyn, F., McAuliffe, F. M. & Wingfield, M. The role of the cervix in fertility: is it time for a reappraisal? Hum. Reprod. 29, 2092–2098 (2014).
WCRF (2018).
Chan, C. K., Aimagambetova, G., Ukybassova, T., Kongrtay, K. & Azizan, A. Human papillomavirus infection and cervical cancer: epidemiology, screening, and vaccination-review of current perspectives. J. Oncol. 2019, 3257939 (2019).
Zhu, H., Shen, Z., Luo, H., Zhang, W. & Zhu, X. Chlamydia trachomatis infection-associated risk of cervical cancer: a meta-analysis. Medicine 95, e3077 (2016).
Shen, R., Richter, H. E. & Smith, P. D. Interactions between HIV-1 and mucosal cells in the female reproductive tract. Am. J. Reprod. Immunol. 71, 608–617 (2014).
WHO (2018).
Schutgens, F. & Clevers, H. Human organoids: tools for understanding biology and treating diseases. Annu. Rev. Pathol. 15, 211–234 (2020).
Kim, J., Koo, B. K. & Knoblich, J. A. Human organoids: model systems for human biology and medicine. Nat. Rev. Mol. Cell Biol. 21, 571–584 (2020).
Koster, S. et al. Modelling Chlamydia and HPV co-infection in patient-derived ectocervix organoids reveals distinct cellular reprogramming. Nat. Commun. 13, 1030 (2022).
Clevers, H., Loh, K. M. & Nusse, R. Stem cell signaling. An integral program for tissue renewal and regeneration: Wnt signaling and stem cell control. Science 346, 1248012 (2014).
Hsu, Y. C., Li, L. & Fuchs, E. Transit-amplifying cells orchestrate stem cell activity and tissue regeneration. Cell 157, 935–949 (2014).
Huch, M. et al. Long-term culture of genome-stable bipotent stem cells from adult human liver. Cell 160, 299–312 (2015).
Rheinwald, J. G. & Green, H. Epidermal growth factor and the multiplication of cultured human epidermal keratinocytes. Nature 265, 421–424 (1977).
Llames, S., García-Pérez, E., Meana, Á., Larcher, F. & del Río, M. Feeder layer cell actions and applications. Tissue Eng. B 21, 345–353 (2015).
Herfs, M. et al. A discrete population of squamocolumnar junction cells implicated in the pathogenesis of cervical cancer. Proc. Natl Acad. Sci. USA 109, 10516–10521 (2012).
Koskela, P. et al. Chlamydia trachomatis infection as a risk factor for invasive cervical cancer. Int. J. Cancer 85, 35–39 (2000).
Ssedyabane, F. et al. HPV-chlamydial coinfection, prevalence, and association with cervical intraepithelial lesions: a pilot study at Mbarara Regional Referral Hospital. J. Cancer Epidemiol. 2019, 9092565 (2019).
Lohmussaar, K. et al. Patient-derived organoids model cervical tissue dynamics and viral oncogenesis in cervical cancer. Cell Stem Cell 28, 1380–1396 (2021).
Hoffmann, K. et al. Stable expansion of high-grade serous ovarian cancer organoids requires a low-Wnt environment. EMBO J. 39, e104013 (2020).
Lohmussaar, K. et al. Assessing the origin of high-grade serous ovarian cancer using CRISPR-modification of mouse organoids. Nat. Commun. 11, 2660 (2020).
Dolat, L. & Valdivia, R.H. An endometrial organoid model of interactions between Chlamydia and epithelial and immune cells. J. Cell Sci. 134, jcs252403 (2021).
Kessler, M. et al. Chronic Chlamydia infection in human organoids increases stemness and promotes age-dependent CpG methylation. Nat. Commun. 10, 1194 (2019).
Fatehullah, A., Tan, S. H. & Barker, N. Organoids as an in vitro model of human development and disease. Nat. Cell Biol. 18, 246–254 (2016).
Sato, T. et al. Single Lgr5 stem cells build crypt–villus structures in vitro without a mesenchymal niche. Nature 459, 262–265 (2009).
Katz, D. F. Human cervical mucus: research update. Am. J. Obstet. Gynecol. 165, 1984–1986 (1991).
Merbah, M. et al. Cervico-vaginal tissue ex vivo as a model to study early events in HIV-1 infection. Am. J. Reprod. Immunol. 65, 268–278 (2011).
Michelini, M., Rosellini, A., Simoncini, T., Papini, S. & Revoltella, R. P. A three-dimensional organotypic culture of the human uterine exocervix for studying mucosal epithelial differentiation and migrating leukocytes. Differentiation 72, 138–149 (2004).
Cattaneo, C. M. et al. Tumor organoid-T-cell coculture systems. Nat. Protoc. 15, 15–39 (2020).
Zadora, P. K. et al. Integrated phosphoproteome and transcriptome analysis reveals chlamydia-induced epithelial-to-mesenchymal transition in host cells. Cell Rep. 26, 1286–1302 (2019).
Farin, H. F., Van, Es,J. H. & Clevers, H. Redundant sources of Wnt regulate intestinal stem cells and promote formation of Paneth cells. Gastroenterology 143, 1518–152s (2012).
Willert, K. et al. Wnt proteins are lipid-modified and can act as stem cell growth factors. Nature 423, 448–452 (2003).
Gurumurthy, R. K. et al. A loss-of-function screen reveals Ras-and Raf-independent MEK–ERK signaling during Chlamydia trachomatis infection. Sci. Signal. 3, ra21 (2010).
Fuerer, C. & Nusse, R. Lentiviral vectors to probe and manipulate the Wnt signaling pathway. PLoS One 5, e9370 (2010).
Acknowledgements
We are thankful to the patients for donating tissue for research. We thank M. Mangler and staff at Vivantes Klinikum Berlin, Charité University Medicine, Berlin, Germany. We thank the late J. Angermann for the technical help with lentivirus production. This work was initiated at the Max Planck Institute for Infection Biology, Berlin. C.C. is funded by the University of Würzburg and DFG (GRK 2157). T.F.M. acknowledges funding from BMBF via the Infect-ERA program CINOCA; N.K. is funded by DFG-DAAD and DFG (GRK2157).
Author information
Authors and Affiliations
Contributions
C.C. and R.K.G. developed human and mouse endocervical and ectocervical organoid culture systems in ref. 4, and T.F.M. provided the infrastructure and guidance. S.K. and N.K. carried out experiments adopting the organoid culture system. S.K., C.C. and R.K.G. developed the genetic manipulation and infection protocols for the organoid system. C.C., R.K.G., S.K. and N.K. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Protocols thanks Yoshitaka Hippo and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key references using this protocol
Chumduri, C. et al. Nat. Cell Biol. 23, 184–197 (2021): https://doi.org/10.1038/s41556-020-00619-0
Koster, S. et al. Nat. Commun. 13, 1030 (2022): https://doi.org/10.1038/s41467-022-28569-1
Source data
Source Data Fig. 3
Unprocessed gels.
Rights and permissions
About this article
Cite this article
Gurumurthy, R.K., Koster, S., Kumar, N. et al. Patient-derived and mouse endo-ectocervical organoid generation, genetic manipulation and applications to model infection. Nat Protoc 17, 1658–1690 (2022). https://doi.org/10.1038/s41596-022-00695-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-022-00695-6
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.