Abstract
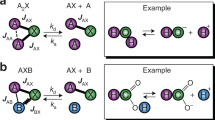
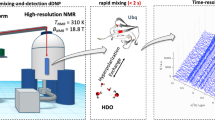
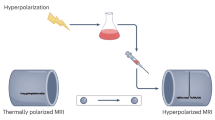
NMR spectroscopy is the only method to access the structural dynamics of biomolecules at high (atomistic) resolution in their native solution state. However, this method’s low sensitivity has two important consequences: (i) typically experiments have to be performed at high concentrations that increase sensitivity but are not physiological, and (ii) signals have to be accumulated over long periods, complicating the determination of interaction kinetics on the order of seconds and impeding studies of unstable systems. Both limitations are of equal, fundamental relevance: non-native conditions are of limited pharmacological relevance, and the function of proteins, enzymes and nucleic acids often relies on their interaction kinetics. To overcome these limitations, we have developed applications that involve ‘hyperpolarized water’ to boost signal intensities in NMR of proteins and nucleic acids. The technique includes four stages: (i) preparation of the biomolecule in partially deuterated buffers, (ii) preparation of ‘hyperpolarized’ water featuring enhanced 1H NMR signals via cryogenic dynamic nuclear polarization, (iii) sudden melting of the cryogenic pellet and dissolution of the protein or nucleic acid in the hyperpolarized water (enabling spontaneous exchanges of protons between water and target) and (iv) recording signal-amplified NMR spectra targeting either labile 1H or neighboring 15N/13C nuclei in the biomolecule. Water in the ensuing experiments is used as a universal ‘hyperpolarization’ agent, rendering the approach versatile and applicable to any biomolecule possessing labile hydrogens. Thus, questions can be addressed, ranging from protein and RNA folding problems to resolving structure-function relationships of intrinsically disordered proteins to investigating membrane interactions.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout









Similar content being viewed by others
Data availability
Data shown in Supplementary Fig. 4 can be found at https://zenodo.org/record/5774664#.YjfH6BDMKqA. All other data are published elsewhere and can be found as referenced in the main text.
References
Ernst, R. R., Bodenhausen, G. & Wokaun, A. Principles of Nuclear Magnetic Resonance in One and Two Dimensions (Clarendon Press, 1987).
Korchak, S. E., Ivanov, K. L., Yurkovskaya, A. V. & Vieth, H. M. Para-hydrogen induced polarization in multi-spin systems studied at variable magnetic field. Phys. Chem. Chem. Phys. 11, 11146–11156 (2009).
Buljubasich, L., Franzoni, M. B., Spiess, H. W. & Munnemann, K. Level anti-crossings in ParaHydrogen Induced Polarization experiments with Cs-symmetric molecules. J. Magn. Reson. 219, 33–40 (2012).
Tokmic, K., Greer, R. B., Zhu, L. & Fout, A. R. 13C NMR signal enhancement using parahydrogen-induced polarization mediated by a cobalt hydrogenation catalyst. J. Am. Chem. Soc. 140, 14844–14850 (2018).
Kiryutin, A. S. et al. Ultrafast single-scan 2D NMR spectroscopic detection of a PHIP-hyperpolarized protease inhibitor. Chemistry 25, 4025–4030 (2019).
Richardson, P. M. et al. Rapid 13C NMR hyperpolarization delivered from para-hydrogen enables the low concentration detection and quantification of sugars. Chem. Sci. 10, 10607–10619 (2019).
Song, B., Choi, D., Xin, Y., Bowers, C. R. & Hagelin-Weaver, H. Ultra-low loading Pt/CeO2 catalysts: ceria facet effect affords improved pairwise selectivity for parahydrogen enhanced NMR spectroscopy. Angew. Chem. Int. Ed. Engl. 60, 4038–4042 (2021).
Raftery, D., MacNamara, E., Fisher, G., Rice, C. V. & Smith, J. Optical pumping and magic angle spinning: sensitivity and resolution enhancement for surface NMR obtained with laser-polarized xenon. J. Am. Chem. Soc. 119, 8746–8747 (1997).
Min, H., Sekar, G. & Hilty, C. Polarization transfer from ligands hyperpolarized by dissolution dynamic nuclear polarization for screening in drug discovery. ChemMedChem 10, 1559–1563 (2015).
Weiland, E., Springuel-Huet, M. A., Nossov, A. & Gedeon, A. 129Xenon NMR: review of recent insights into porous materials. Microporous Mesoporous Mater. 225, 41–65 (2016).
Khan, A. S. et al. Enabling clinical technologies for hyperpolarized 129xenon magnetic resonance imaging and spectroscopy. Angew. Chem. Int. Ed. Engl. 60, 22126–22147 (2021).
King, J. P. et al. Room-temperature in situ nuclear spin hyperpolarization from optically pumped nitrogen vacancy centres in diamond. Nat. Commun. 6, 8965 (2015).
Alvarez, G. A. et al. Local and bulk 13C hyperpolarization in nitrogen-vacancy-centred diamonds at variable fields and orientations. Nat. Commun. 6, 8456 (2015).
Fernandez-Acebal, P. et al. Toward hyperpolarization of oil molecules via single nitrogen vacancy centers in diamond. Nano Lett. 18, 1882–1887 (2018).
Ajoy, A. et al. Orientation-independent room temperature optical 13C hyperpolarization in powdered diamond. Sci. Adv. 4, eaar5492 (2018).
Tateishi, K. et al. Room temperature hyperpolarization of nuclear spins in bulk. Proc. Natl Acad. Sci. USA 111, 7527–7530 (2014).
Kouno, H. et al. Triplet dynamic nuclear polarization of crystalline ice using water-soluble polarizing agents. Chem. Commun. (Camb.) 56, 3717–3720 (2020).
Armstrong, B. D. & Han, S. Overhauser dynamic nuclear polarization to study local water dynamics. J. Am. Chem. Soc. 131, 4641–4647 (2009).
Neugebauer, P. et al. Liquid state DNP of water at 9.2 T: an experimental access to saturation. Phys. Chem. Chem. Phys. 15, 6049–6056 (2013).
Can, T. V., Ni, Q. Z. & Griffin, R. G. Mechanisms of dynamic nuclear polarization in insulating solids. J. Magn. Reson. 253, 23–35 (2015).
Pylaeva, S., Ivanov, K. L., Baldus, M., Sebastiani, D. & Elgabarty, H. Molecular mechanism of Overhauser dynamic nuclear polarization in insulating solids. J. Phys. Chem. Lett. 8, 2137–2142 (2017).
Wang, Y. & Hilty, C. Amplification of nuclear Overhauser effect signals by hyperpolarization for screening of ligand binding to immobilized target proteins. Anal. Chem. 92, 13718–13723 (2020).
Kircher, R., Hasse, H. & Munnemann, K. High flow-rate benchtop NMR spectroscopy enabled by continuous Overhauser DNP. Anal. Chem. 93, 8897–8905 (2021).
Liu, G. et al. One-thousand-fold enhancement of high field liquid nuclear magnetic resonance signals at room temperature. Nat. Chem. 9, 676–680 (2017).
Loening, N. M., Rosay, M., Weis, V. & Griffin, R. G. Solution-state dynamic nuclear polarization at high magnetic field. J. Am. Chem. Soc. 124, 8808 (2002).
Hofer, P. et al. Field dependent dynamic nuclear polarization with radicals in aqueous solution. J. Am. Chem. Soc. 130, 3254 (2008).
Dubroca, T., Wi, S., van Tol, J., Frydman, L. & Hill, S. Large volume liquid state scalar Overhauser dynamic nuclear polarization at high magnetic field. Phys. Chem. Chem. Physi. 21, 21200 (2019).
Neugebauer, P. et al. Liquid state DNP of water at 9.2 T: an experimental access to saturation. Phys. Chem. Chem. Phys. 15, 6049–6056 (2013).
van Bentum, J., van Meerten, B., Sharma, M. & Kentgens, A. Perspectives on DNP-enhanced NMR spectroscopy in solutions. J. Magn. Reson. 264, 59–67 (2016).
Abragam, A. & Goldman, M. Principles of dynamic nuclear polarisation. Rep. Prog. Phys. 41, 395–467 (1978).
Shimon, D., Hovav, Y., Feintuch, A., Goldfarb, D. & Vega, S. Dynamic nuclear polarization in the solid state: a transition between the cross effect and the solid effect. Phys. Chem. Chem. Phys. 14, 5729–5743 (2012).
Borghini, M., Deboer, W. & Morimoto, K. Nuclear dynamic polarization by resolved solid-state effect and thermal mixing with an electron spin-spin interaction reservoir. Phys. Lett. A 48, 244–246 (1974).
Henstra, A. & Wenckebach, W. T. Dynamic nuclear polarisation via the integrated solid effect I: theory. Mol. Phys. 112, 1761–1772 (2014).
Wenckebach, W. T. Dynamic nuclear polarization via thermal mixing: beyond the high temperature approximation. J. Magn. Reson. 277, 68–78 (2017).
Wenckebach, W. T. Spectral diffusion and dynamic nuclear polarization: beyond the high temperature approximation. J. Magn. Reson. 284, 104–114 (2017).
Wenckebach, W. T. Dynamic nuclear polarization via the cross effect and thermal mixing: B. Energy transport. J. Magn. Reson. 299, 151–167 (2019).
Ardenkjaer-Larsen, J. H. et al. Increase in signal-to-noise ratio of > 10,000 times in liquid-state NMR. Proc. Natl Acad. Sci. USA 100, 10158–10163 (2003).
Golman, K., Ardenaer-Larsen, J. H., Petersson, J. S., Mansson, S. & Leunbach, I. Molecular imaging with endogenous substances. Proc. Natl Acad. Sci. USA 100, 10435–10439 (2003).
Jannin, S., Dumez, J. N., Giraudeau, P. & Kurzbach, D. Application and methodology of dissolution dynamic nuclear polarization in physical, chemical and biological contexts. J. Magn. Reson. 305, 41–50 (2019).
Rosay, M. et al. Solid-state dynamic nuclear polarization at 263 GHz: spectrometer design and experimental results. Phys. Chem. Chem. Phys. 12, 5850–5860 (2010).
Akbey, U. & Oschkinat, H. Structural biology applications of solid state MAS DNP NMR. J. Magn. Reson. 269, 213–224 (2016).
Kaplan, M. et al. Probing a cell-embedded megadalton protein complex by DNP-supported solid-state NMR. Nat. Methods 12, 649–652 (2015).
Carnevale, D. et al. Natural abundance oxygen-17 solid-state NMR of metal organic frameworks enhanced by dynamic nuclear polarization. Phys. Chem. Chem. Phys. 23, 2245–2251 (2021).
Pinon, A. C., Rossini, A. J., Widdifield, C. M., Gajan, D. & Emsley, L. Polymorphs of theophylline characterized by DNP enhanced solid-state NMR. Mol. Pharm. 12, 4146–4153 (2015).
Lesage, A. et al. Surface enhanced NMR spectroscopy by dynamic nuclear polarization. J. Am. Chem. Soc. 132, 15459–15461 (2010).
Harris, T., Szekely, O. & Frydman, L. On the potential of hyperpolarized water in biomolecular NMR studies. J. Phys. Chem. B 118, 3281–3290 (2014).
Giraudeau, P., Muller, N., Jerschow, A. & Frydman, L. H-1 NMR noise measurements in hyperpolarized liquid samples. Chem. Phys. Lett. 489, 107–112 (2010).
Leftin, A., Roussel, T. & Frydman, L. Hyperpolarized functional magnetic resonance of murine skeletal muscle enabled by multiple tracer-paradigm synchronizations. Plos One 9, e96399 (2014).
Frydman, L. & Blazina, D. Ultrafast two-dimensional nuclear magnetic resonance spectroscopy of hyperpolarized solutions. Nat. Phys. 3, 415–419 (2007).
Jeschke, G. & Frydman, L. Nuclear hyperpolarization comes of age. J. Magn. Reson. 264, 1–2 (2016).
Leftin, A., Degani, H. & Frydman, L. In vivo magnetic resonance of hyperpolarized [13C1]pyruvate: metabolic dynamics in stimulated muscle. Am. J. Physiol. Endocrinol. Metab. 305, E1165–E1171 (2013).
Markovic, S. et al. Placental physiology monitored by hyperpolarized dynamic 13C magnetic resonance. Proc. Natl Acad. Sci. USA 115, E2429–E2436 (2018).
Hwang, J.-H. & Choi, C. S. Use of in vivo magnetic resonance spectroscopy for studying metabolic diseases. Exp. Mol. Med. 47, e139 (2015).
Can, T. V. et al. Overhauser effects in insulating solids. J. Chem. Phys. 141, 064202 (2014).
Maly, T. et al. Dynamic nuclear polarization at high magnetic fields. J. Chem. Phys. 128, 052211 (2008).
Can, T. V. et al. Frequency-swept integrated and stretched solid effect dynamic nuclear polarization. J. Phys. Chem. Lett. 9, 3187–3192 (2018).
Mentink-Vigier, F. Optimizing nitroxide biradicals for cross-effect MAS-DNP: the role of g-tensors’ distance. Phys. Chem. Chem. Phys. 22, 3643–3652 (2020).
Harris, T., Eliyahu, G., Frydman, L. & Degani, H. Kinetics of hyperpolarized 13C1-pyruvate transport and metabolism in living human breast cancer cells. Proc. Natl Acad. Sci. USA 106, 18131–18136 (2009).
Chen, A. P. et al. Hyperpolarized C-13 spectroscopic imaging of the TRAMP mouse at 3T—initial experience. Magn. Reson. Med. 58, 1099–1106 (2007).
Gallagher, F. A. et al. Magnetic resonance imaging of pH in vivo using hyperpolarized 13C-labelled bicarbonate. Nature 453, 940–943 (2008).
Lesage, A. et al. Surface enhanced NMR spectroscopy by dynamic nuclear polarization. J. Am. Chem. Soc. 132, 15459–15461 (2010).
Thankamony, A. S. L., Wittmann, J. J., Kaushik, M. & Corzilius, B. Dynamic nuclear polarization for sensitivity enhancement in modern solid-state NMR. Prog. Nucl. Magn. Reson. Spectrosc. 102, 120–195 (2017).
Jan Henrik Ardenkjær-Larsen, eMagRes. 7, 63–78 (2018).
Tan, K. O., Yang, C., Weber, R. T., Mathies, G. & Griffin, R. G. Time-optimized pulsed dynamic nuclear polarization. Sci. Adv. 5, eaav6909 (2019).
Capozzi, A. et al. Efficient hyperpolarization of U-13C-glucose using narrow-line UV-generated labile free radicals. Angew. Chem. Int. Ed. Engl. 58, 1334–1339 (2019).
Pinon, A. C., Capozzi, A. & Ardenkjær-Larsen, J. H. Hyperpolarized water through dissolution dynamic nuclear polarization with UV-generated radicals. Commun. Chem. 3, 57 (2020).
Eichhorn, T. R. et al. Hyperpolarization without persistent radicals for in vivo real-time metabolic imaging. Proc. Natl Acad. Sci. USA 110, 18064–18069 (2013).
Capozzi, A., Cheng, T., Boero, G., Roussel, C. & Comment, A. Thermal annihilation of photo-induced radicals following dynamic nuclear polarization to produce transportable frozen hyperpolarized 13C-substrates. Nat. Commun. 8, 15757 (2017).
Gajan, D. et al. Hybrid polarizing solids for pure hyperpolarized liquids through dissolution dynamic nuclear polarization. Proc. Natl Acad. Sci. USA 111, 14693–14697 (2014).
Baudouin, D. et al. Cubic three-dimensional hybrid silica solids for nuclear hyperpolarization. Chem. Sci. 7, 6846–6850 (2016).
Katz, I. & Blank, A. Dynamic nuclear polarization in solid samples by electrical-discharge-induced radicals. J. Magn. Reson. 261, 95–100 (2015).
Katz, I., Feintuch, A., Carmieli, R. & Blank, A. Proton polarization enhancement of up to 150 with dynamic nuclear polarization of plasma-treated glucose powder. Solid State Nucl. Magn. Reson. 100, 26–35 (2019).
Blank, A., Katz, I. & A. Feintuch. Method for Preparation of Highly Polarized Nuclear Spins Containing Samples and Uses Thereof for NMR and MRI. USA patent 10,718,840 (2020).
Carnahan, S. L., Venkatesh, A., Perras, F. A., Wishart, J. F. & Rossini, A. J. High-field magic angle spinning dynamic nuclear polarization using radicals created by γ-irradiation. J. Phys. Chem. Lett. 10, 4770–4776 (2019).
Kouřil, K., Kouřilová, H., Bartram, S., Levitt, M. H. & Meier, B. Scalable dissolution-dynamic nuclear polarization with rapid transfer of a polarized solid. Nat. Commun. 10, 1733 (2019).
van Meerten, S. G. J., Janssen, G. E. & Kentgens, A. P. M. Rapid-melt DNP for multidimensional and heteronuclear high-field NMR experiments. J. Magn. Reson. 310, 106656 (2020).
van Bentum, P. J. M., Sharma, M., van Meerten, S. G. J. & Kentgens, A. P. M. Solid effect DNP in a rapid-melt setup. J. Magn. Reson. 263, 126–135 (2016).
Joo, C. G., Hu, K. N., Bryant, J. A. & Griffin, R. G. In situ temperature jump high-frequency dynamic nuclear polarization experiments: enhanced sensitivity in liquid-state NMR spectroscopy. J. Am. Chem. Soc. 128, 9428–9432 (2006).
Yoon, D. et al. 500-fold enhancement of in situ 13C liquid state NMR using gyrotron-driven temperature-jump DNP. J. Magn. Reson. 270, 142–146 (2016).
Hovav, Y., Feintuch, A. & Vega, S. Theoretical aspects of dynamic nuclear polarization in the solid state — the cross effect. J. Magn. Reson. 214, 29–41 (2012).
Leavesley, A. et al. Effect of electron spectral diffusion on static dynamic nuclear polarization at 7 Tesla. Phys. Chem. Chem. Phys. 19, 3596–3605 (2017).
Ji, X. et al. Overhauser effects in non-conducting solids at 1.2K. J. Magn. Reson. 286, 138–142 (2017).
Jannin, S., Comment, A. & van der Klink, J. J. Dynamic nuclear polarization by thermal mixing under partial saturation. Appl. Magn. Reson. 43, 59–68 (2012).
Wenckebach, W. T. Dynamic nuclear polarization via thermal mixing: beyond the high temperature approximation. J. Magn. Reson. 277, 68–78 (2017).
Tayler, M. C. et al. Direct enhancement of nuclear singlet order by dynamic nuclear polarization. J. Am. Chem. Soc. 134, 7668–7671 (2012).
Miclet, E. et al. Toward quantitative measurements of enzyme kinetics by dissolution dynamic nuclear polarization. J. Phys. Chem. Lett. 5, 3290–3295 (2014).
Buratto, R. et al. Drug screening boosted by hyperpolarized long-lived states in NMR. ChemMedChem 9, 2509–2515 (2014).
Mammoli, D. et al. Hyperpolarized para-ethanol. J. Phys. Chem. B 119, 4048–4052 (2015).
Dumez, J. N. et al. Hyperpolarized NMR of plant and cancer cell extracts at natural abundance. Analyst 140, 5860–5863 (2015).
Ragavan, M., Iconaru, L. I., Park, C. G., Kriwacki, R. W. & Hilty, C. Real-time analysis of folding upon binding of a disordered protein by using dissolution DNP NMR spectroscopy. Angew. Chem. Int. Ed. Engl. 56, 7070–7073 (2017).
Ji, X. et al. Transportable hyperpolarized metabolites. Nat. Commun. 8, 13975 (2017).
Nardi-Schreiber, A. et al. Biochemical phosphates observed using hyperpolarized 31P in physiological aqueous solutions. Nat. Commun. 8, 341 (2017).
Kovtunov, K. V. et al. Hyperpolarized NMR spectroscopy: d-DNP, PHIP, and SABRE techniques. Chem. Asian J. In press. https://doi.org/10.1002/asia.201800551 (2018).
Weber, E. M. M. et al. Assessing the onset of calcium phosphate nucleation by hyperpolarized real-time NMR. Anal. Chem. 92, 7666–7673 (2020).
Ardenkjaer-Larsen, J. H. et al. Cryogen-free dissolution dynamic nuclear polarization polarizer operating at 3.35 T, 6.70 T, and 10.1 T. Magn. Reson. Med. 81, 2184–2194 (2019).
Kim, J., Mandal, R. & Hilty, C. Characterization of membrane protein-lipid interactions in unfolded OmpX with enhanced time resolution by hyperpolarized NMR. Chembiochem 21, 2861–2867 (2020).
Bloembergen, N., Purcell, E. M. & Pound, R. V. Nuclear magnetic relaxation. Nature 160, 475–476 (1947).
Bloembergen, N., Purcell, E. M. & Pound, R. V. Relaxation effects in nuclear magnetic resonance Absorption. Phys. Rev. 73, 679 (1948).
Honegger, P. & Steinhauser, O. The protein-water nuclear Overhauser effect (NOE) as an indirect microscope for molecular surface mapping of interaction patterns. Phys. Chem. Chem. Phys. 22, 212–222 (2019).
Reese, M. et al. 1H and 13C dynamic nuclear polarization in aqueous solution with a two-field (0.35 T/14 T) shuttle DNP spectrometer. J. Am. Chem. Soc. 131, 15086–15087 (2009).
Doll, A., Bordignon, E., Joseph, B., Tschaggelar, R. & Jeschke, G. Liquid state DNP for water accessibility measurements on spin-labeled membrane proteins at physiological temperatures. J. Magn. Reson. 222, 34–43 (2012).
Chen, H. Y., Ragavan, M. & Hilty, C. Protein folding studied by dissolution dynamic nuclear polarization. Angew. Chem. Int. Ed. Engl. 52, 9192–9195 (2013).
Wang, Y., Kim, J. & Hilty, C. Determination of protein-ligand binding modes using fast multi-dimensional NMR with hyperpolarization. Chem. Sci. 11, 5935–5943 (2020).
Qi, C., Wang, Y. & Hilty, C. Application of relaxation dispersion of hyperpolarized 13 C spins to protein-ligand binding. Angew. Chem. Int. Ed. Engl. 60, 24018–24021 (2021).
Harris, T., Szekely, O. & Frydman, L. On the potential of hyperpolarized water in biomolecular NMR studies. J. Phys. Chem. B 118, 3281–3290 (2014).
Olsen, G., Markhasin, E., Szekely, O., Bretschneider, C. & Frydman, L. Optimizing water hyperpolarization and dissolution for sensitivity-enhanced 2D biomolecular NMR. J. Magn. Reson. 264, 49–58 (2016).
Kurzbach, D. et al. Investigation of intrinsically disordered proteins through exchange with hyperpolarized water. Angew. Chem. Int. Ed. Engl. 56, 389–392 (2017).
Kim, J., Liu, M. & Hilty, C. Modeling of polarization transfer kinetics in protein hydration using hyperpolarized water. J. Phys. Chem. B 121, 6492–6498 (2017).
Szekely, O., Olsen, G. L., Felli, I. C. & Frydman, L. High-resolution 2D NMR of disordered proteins enhanced by hyperpolarized water. Anal. Chem. 90, 6169–6177 (2018).
Sadet, A. et al. Hyperpolarized water enhances two-dimensional proton NMR correlations: a new approach for molecular interactions. J. Am. Chem. Soc. 141, 12448–12452 (2019).
Olsen, G. L. et al. Sensitivity-enhanced three-dimensional and carbon-detected two-dimensional NMR of proteins using hyperpolarized water. J. Biomol. NMR 74, 161–171 (2020).
Szekely, O., Olsen, G. L., Novakovic, M., Rosenzweig, R. & Frydman, L. Assessing site-specific enhancements imparted by hyperpolarized water in folded and unfolded proteins by 2D HMQC NMR. J. Am. Chem. Soc. 142, 9267–9284 (2020).
Novakovic, M. et al. A 300-fold enhancement of imino nucleic acid resonances by hyperpolarized water provides a new window for probing RNA refolding by 1D and 2D NMR. Proc. Natl Acad. Sci. USA 117, 2449–2455 (2020).
Kim, J., Mandal, R. & Hilty, C. 2D NMR spectroscopy of refolding RNase Sa using polarization transfer from hyperpolarized water. J. Magn. Reson. 326, 106942 (2021).
Kadeřávek, P., Ferrage, F., Bodenhausen, G. & Kurzbach, D. High-resolution NMR of folded proteins in hyperpolarized physiological solvents. Chemistry 24, 13418–13423 (2018).
Liu, M. & Hilty, C. Metabolic measurements of nonpermeating compounds in live cells using hyperpolarized NMR. Anal. Chem. 90, 1217–1222 (2018).
Lehmkuhl, S. et al. Continuous hyperpolarization with parahydrogen in a membrane reactor. J. Magn. Reson. 291, 8–13 (2018).
Krajewski, M. et al. A multisample dissolution dynamic nuclear polarization system for serial injections in small animals. Magn. Reson. Med. 77, 904–910 (2017).
Wilson, D. M. & Kurhanewicz, J. Hyperpolarized 13C MR for molecular imaging of prostate cancer. J. Nucl. Med. 55, 1567–1572 (2014).
Nikolaou, P., Goodson, B. M. & Chekmenev, E. Y. NMR hyperpolarization techniques for biomedicine. Chemistry 21, 3156–3166 (2015).
Milani, J. et al. A magnetic tunnel to shelter hyperpolarized fluids. Rev. Sci. Instrum. 86, 024101 (2015).
Chen, H. Y. & Hilty, C. Implementation and characterization of flow injection in dissolution dynamic nuclear polarization NMR spectroscopy. ChemPhysChem 16, 2646–2652 (2015).
Katsikis, S., Marin-Montesinos, I., Pons, M., Ludwig, C. & Gunther, U. L. Improved stability and spectral quality in ex situ dissolution DNP using an improved transfer device. Appl. Magn. Reson. 46, 723–729 (2015).
Bowen, S. & Hilty, C. Rapid sample injection for hyperpolarized NMR spectroscopy. Phys. Chem. Chem. Phys. 12, 5766–5770 (2010).
Vuichoud, B. et al. Filterable agents for hyperpolarization of water, metabolites, and proteins. Chem. Eur. J. 22, 14696–14700 (2016).
Bodenhausen, G. Heteronuclear J-spectroscopy. J. Magn. Reson. 39, 175–179 (1980).
Negroni, M. & Kurzbach, D. Residue-resolved monitoring of protein hyperpolarization at sub-second time resolution. Commun. Chem. 4, 127 (2021).
Chappuis, Q. et al. Hyperpolarized water to study protein-ligand interactions. J. Phys. Chem. Lett. 6, 1674–1678 (2015).
Hwanga, T.-L., van Zijl, P. C. & Mori, S. Accurate quantitation of water–amide proton exchange rates using the Phase-Modulated CLEAN chemical EXchange (CLEANEX-PM) approach with a Fast-HSQC (FHSQC) detection scheme. J. Biomol. Nmr. 11, 221–226 (1998).
Nucci, N. V., Pometun, M. S. & Wand, A. J. Site-resolved measurement of water-protein interactions by solution NMR. Nat. Struct. Mol. Biol. 18, 245–249 (2011).
Schanda, P. & Brutscher, B. Hadamard frequency-encoded SOFAST-HMQC for ultrafast two-dimensional protein NMR. J. Magn. Reson. 178, 334–339 (2006).
Mazhab-Jafari, M. T. et al. Understanding cAMP-dependent allostery by NMR spectroscopy: comparative analysis of the EPAC1 cAMP-binding domain in its apo and cAMP-bound states. J. Am. Chem. Soc. 129, 14482–14492 (2007).
Dalvit, C., Fogliatto, G., Stewart, A., Veronesi, M. & Stockman, B. WaterLOGSY as a method for primary NMR screening: practical aspects and range of applicability. J. Biomol. NMR 21, 349–359 (2001).
Frieden, C., Hoeltzli, S. D. & Ropson, I. J. NMR and protein folding: equilibrium and stopped-flow studies. Protein Sci. 2, 2007–2014 (1993).
Charlier, C. et al. Study of protein folding under native conditions by rapidly switching the hydrostatic pressure inside an NMR sample cell. Proc. Natl Acad. Sci. USA 115, E4169–E4178 (2018).
Dobson, C. M. & Hore, P. J. Kinetic studies of protein folding using NMR spectroscopy. Nat. Struct. Biol. 5(Suppl), 504–507 (1998).
Krahn, A. et al. Shuttle DNP spectrometer with a two-center magnet. Phys. Chem. Chem. Phys. 12, 5830–5840 (2010).
Franck, J. M., Ding, Y., Stone, K., Qin, P. Z. & Han, S. Anomalously rapid hydration water diffusion dynamics near DNA surfaces. J. Am. Chem. Soc. 137, 12013–12023 (2015).
Fisette, O. et al. Hydration dynamics of a peripheral membrane protein. J. Am. Chem. Soc. 138, 11526–11535 (2016).
Armstrong, B. D. et al. Site-specific hydration dynamics in the nonpolar core of a molten globule by dynamic nuclear polarization of water. J. Am. Chem. Soc. 133, 5987–5995 (2011).
Pavlova, A. et al. Site-specific dynamic nuclear polarization of hydration water as a generally applicable approach to monitor protein aggregation. Phys. Chem. Chem. Phys. 11, 6833–6839 (2009).
Yang, W. Y. & Gruebele, M. Folding at the speed limit. Nature 423, 193–197 (2003).
Phillips, J. C., LeGrand, A. D. & Lehnert, W. F. Protein folding observed by time-resolved synchrotron x-ray scattering. A feasibility study. Biophys. J. 53, 461–464 (1988).
Ecevit, O., Khan, M. A. & Goss, D. J. Kinetic analysis of the interaction of b/HLH/Z transcription factors Myc, Max, and Mad with cognate DNA. Biochemistry 49, 2627–2635 (2010).
Josts, I. et al. Structural kinetics of MsbA investigated by stopped-flow time-resolved small-angle X-ray scattering. Structure 28, 348–354.e3 (2020).
Vancraenenbroeck, R. & Hofmann, H. Occupancies in the DNA-binding pathways of intrinsically disordered helix-loop-helix leucine-zipper proteins. J. Phys. Chem. B 122, 11460–11467 (2018).
Kim, Y., Liu, M. X. & Hilty, C. Parallelized ligand screening using dissolution dynamic nuclear polarization. Anal. Chem. 88, 11178–11183 (2016).
Kurzbach, D. et al. Dissolution dynamic nuclear polarization of deuterated molecules enhanced by cross-polarization. J. Chem. Phys. 145, 194203 (2016).
Bowen, S. & Hilty, C. Time-resolved dynamic nuclear polarization enhanced NMR spectroscopy. Angew. Chem. Int. Ed. Engl. 47, 5235–5237 (2008).
Jannin, S. et al. A 140 GHz prepolarizer for dissolution dynamic nuclear polarization. J. Chem. Phys. 128, 241102 (2008).
Kouřil, K., Kouřilová, H., Levitt, M. H. & Meier, B. Scalable dissolution-dynamic nuclear polarization with rapid transfer of a polarized solid. Natue Communications 10, 1733 (2019).
Kim, J., Mandal, R. & Hilty, C. Observation of fast two-dimensional NMR spectra during protein folding using polarization transfer from hyperpolarized water. J. Phys. Chem. Lett. 10, 5463–5467 (2019).
Lescop, E., Schanda, P. & Brutscher, B. A set of BEST triple-resonance experiments for time-optimized protein resonance assignment. J. Magn. Reson. 187, 163–169 (2007).
Rule, G. S. & Hitchens, T. K. Fundamentals of Protein NMR Spectroscopy (Springer, 2006).
Gil, S. et al. NMR spectroscopic studies of intrinsically disordered proteins at near-physiological conditions. Angew. Chem. Int. Ed. Engl. 52, 11808–11812 (2013).
Kazimierczuk, K., Zawadzka-Kazimierczuk, A. & Kozminski, W. Non-uniform frequency domain for optimal exploitation of non-uniform sampling. J. Magn. Reson. 205, 286–292 (2010).
Kupce, E. & Freeman, R. Wideband excitation with polychromatic pulses. J. Magn. Reson. A 108, 268–273 (1994).
Geen, H. & Freeman, R. Band-selective radiofrequency pulses. J. Magn. Reson. 93, 93–141 (1991).
Hyberts, S. G., Milbradt, A. G., Wagner, A. B., Arthanari, H. & Wagner, G. Application of iterative soft thresholding for fast reconstruction of NMR data non-uniformly sampled with multidimensional Poisson Gap scheduling. J. Biomol. NMR 52, 315–327 (2012).
Kim, J., Liu, M., Chen, H. Y. & Hilty, C. Determination of intermolecular interactions using polarization compensated heteronuclear Overhauser effect of hyperpolarized spins. Anal. Chem. 87, 10982–10987 (2015).
Lee, Y., Zeng, H., Ruedisser, S., Gossert, A. D. & Hilty, C. Nuclear magnetic resonance of hyperpolarized fluorine for characterization of protein-ligand interactions. J. Am. Chem. Soc. 134, 17448–17451 (2012).
Day, I. J., Mitchell, J. C., Snowden, M. J. & Davis, A. L. Applications of DNP-NMR for the measurement of heteronuclear T1 relaxation times. J. Magn. Reson. 187, 216–224 (2007).
Kiryutin, A. S. et al. Transport of hyperpolarized samples in dissolution-DNP experiments. Phys. Chem. Chem. Phys. 21, 13696–13705 (2019).
Mieville, P., Jannin, S. & Bodenhausen, G. Relaxometry of insensitive nuclei: optimizing dissolution dynamic nuclear polarization. J. Magn. Reson. 210, 137–140 (2011).
Kress, T. et al. A novel sample handling system for dissolution dynamic nuclear polarization experiments. Magn. Reson. 2, 387–394 (2021).
Dey, A. et al. Hyperpolarized NMR metabolomics at natural 13C abundance. Anal. Chem. 92, 14867–14871 (2020).
Baudin, M., Vuichoud, B., Bornet, A., Bodenhausen, G. & Jannin, S. A cryogen-consumption-free system for dynamic nuclear polarization at 9.4 T. J. Magn. Reson. 294, 115–121 (2018).
Kurzbach, D., Yao, S., Hinderberger, D. & Klinkhammer, K. W. EPR spectroscopic characterization of persistent germyl-substituted Pb(III)- and Sn(III)-radicals. Dalton Trans. 39, 6449–6459 (2010).
Rangaswami, H., Bulbule, A. & Kundu, G. C. Osteopontin: role in cell signaling and cancer progression. Trends Cell Biol. 16, 79–87 (2006).
Rodrigues, L. R., Teixeira, J. A., Schmitt, F. L., Paulsson, M. & Lindmark-Mansson, H. The role of osteopontin in tumor progression and metastasis in breast cancer. Cancer Epidemiol. Biomark. Prev. 16, 1087–1097 (2007).
Platzer, G. et al. The metastasis-associated extracellular matrix protein osteopontin forms transient structure in ligand interaction sites. Biochemistry 50, 6113–6124 (2011).
Kurzbach, D. et al. Cooperative unfolding of compact conformations of the intrinsically disordered protein osteopontin. Biochemistry 52, 5167–5175 (2013).
Kurzbach, D. et al. Compensatory adaptations of structural dynamics in an intrinsically disordered protein complex. Angew. Chem. Int. Ed. Engl. 53, 3840–3843 (2014).
Acknowledgements
L.F. thanks G. Olsen, O. Szekely, M. Novakovic, K. Singh and C. Bretschneider, who contributed to his training and understanding of events involved in HyperW NMR. Research at the Weizmann Institute of Science is supported by the German-Israel Foundation (grant G-1501-302), the EU Horizon 2020 program (FET-OPEN grant 828946, PATHOS), Israel Science Foundation grant 965/18 and the Perlman Family Foundation. L.F. holds the Bertha and Isadore Gudelsky Professorial Chair and heads the Clore Institute for High-Field Magnetic Resonance Imaging and Spectroscopy, whose support is also acknowledged. C.H. acknowledges support from the National Institutes of Health (grant R01GM132655), the National Science Foundation (grant CHE-1362691) and the Welch Foundation (grant A-1658). D.K. acknowledges contributions from E. Canet, P. Kadeřávek, G. Olsen, D. Guarin and E. M. M. Weber and thanks G. Bodenhausen, F. Ferrage and D. Abergel for their support. The project leading to this application at the University of Vienna received funding from the European Research Council under the EU Horizon 2020 research and innovation programme (grant agreement 801936). Furthermore, this project was supported by the Austrian FWF (standalone grant no. P-33338).
Author information
Authors and Affiliations
Contributions
D.K. initiated and organized the collaboration leading to this Article. All authors wrote and approved the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Protocols thanks Nandita Abhyankar and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related Links
Key references using this protocol
Novakovic, M. et al. Proc. Natl. Acad. Sci. USA 117, 2449–2455 (2020): https://www.pnas.org/content/117/5/2449
Kim, J. et al. J. Phys. Chem. Lett. 10, 5463–5467 (2019): https://doi.org/10.1021/acs.jpclett.9b02197
Szekely, O. et al. J. Am. Chem. Soc. 142, 9267–9284 (2020): https://doi.org/10.1021/jacs.0c00807
Kurzbach, D. et al. Angew. Chem. Int. Ed. Engl. 56, 389–392 (2017): https://doi.org/10.1002/anie.201608903
Sadet, A. et al. J. Am. Chem. Soc. 141, 12448–12452 (2019): https://doi.org/10.1021/jacs.9b03651
Kadeřávek, P. et al. Chemistry 24, 13418–13423 (2018): https://doi.org/10.1002/chem.201802885
Key data used in this protocol
Novakovic, M. et al. Proc. Natl. Acad. Sci. USA 117, 2449–2455 (2020): https://www.pnas.org/content/117/5/2449
Kim, J. et al. J. Magn. Reson. 326, 106942 (2021): https://doi.org/10.1016/j.jmr.2021.106942
Szekely, O. et al. J. Am. Chem. Soc. 142, 9267–9284 (2020): https://doi.org/10.1021/jacs.0c00807
Kurzbach, D. et al. Angew. Chem. Int. Ed. Engl. 56, 389–392 (2017): https://doi.org/10.1002/anie.201608903
Sadet, A. et al. J. Am. Chem. Soc. 141, 12448–12452 (2019): https://doi.org/10.1021/jacs.9b03651
Kadeřávek, P. et al. Chemistry 24, 13418–13423 (2018): https://doi.org/10.1002/chem.201802885
Supplementary information
Supplementary Information
Supplementary Figs. 1–5
Rights and permissions
About this article
Cite this article
Hilty, C., Kurzbach, D. & Frydman, L. Hyperpolarized water as universal sensitivity booster in biomolecular NMR. Nat Protoc 17, 1621–1657 (2022). https://doi.org/10.1038/s41596-022-00693-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-022-00693-8
This article is cited by
-
SHARPER-DOSY: Sensitivity enhanced diffusion-ordered NMR spectroscopy
Nature Communications (2023)
-
Multinuclear 1D and 2D NMR with 19F-Photo-CIDNP hyperpolarization in a microfluidic chip with untuned microcoil
Nature Communications (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.