Abstract
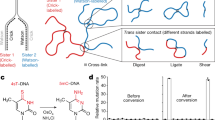
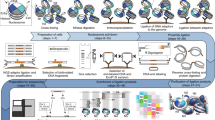
Chromosome conformation capture (Hi-C) techniques map the 3D organization of entire genomes. How sister chromatids fold in replicated chromosomes, however, cannot be determined with conventional Hi-C because of the identical DNA sequences of sister chromatids. Here, we present a protocol for sister chromatid–sensitive Hi-C (scsHi-C) that enables the distinction of DNA contacts within individual sister chromatids (cis sister contacts) from those between sister chromatids (trans sister contacts), thereby allowing investigation of the organization of replicated genomes. scsHi-C is based on live-cell labeling of nascent DNA by the synthetic nucleoside 4-thio-thymidine (4sT), which incorporates into a distinct DNA strand on each sister chromatid because of semi-conservative DNA replication. After purification of genomic DNA and in situ Hi-C library preparation, 4sT is chemically converted into 5-methyl-cytosine in the presence of OsO4/NH4Cl to introduce T-to-C signature point mutations on 4sT-labeled DNA. The Hi-C library is then sequenced, and ligated fragments are assigned to sister chromatids on the basis of strand orientation and the presence of signature mutations. The ensemble of scsHi-C contacts thereby represents genome-wide contact probabilities within and across sister chromatids. scsHi-C can be completed in 2 weeks, has been successfully applied in HeLa cells and can potentially be established for any cell type that allows proper cell cycle synchronization and incorporation of sufficient amounts of 4sT. The genome-wide maps of replicated chromosomes detected by scsHi-C enable investigation of the molecular mechanisms shaping sister chromatid topologies and the relevance of sister chromatid conformation in crucial processes like DNA repair, mitotic chromosome formation and potentially other biological processes.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Code availability
The code used to generate the figures in this manuscript was originally published in Mitter et al.33. Specifically, the ipython notebooks to generate all the plots shown in this manuscript can be found at https://github.com/gerlichlab/scshic_analysis78. A programming environment to perform all analyses shown within this manuscript is provided as a docker container at https://hub.docker.com/repository/docker/gerlichlab/scshic_docker71. The preprocessing pipeline that can be used to convert raw data to .mcool files is available at https://github.com/gerlichlab/scshic_pipeline72.
References
Dekker, J. & Mirny, L. The 3D genome as moderator of chromosomal communication. Cell 164, 1110–1121 (2016).
Rowley, M. J. & Corces, V. G. Organizational principles of 3D genome architecture. Nat. Rev. Genet. 19, 789–800 (2018).
Finn, E. H. & Misteli, T. Molecular basis and biological function of variability in spatial genome organization. Science 365, eaaw9498 (2019).
Davidson, I. F. & Peters, J.-M. Genome folding through loop extrusion by SMC complexes. Nat. Rev. Mol. Cell Biol. 22, 445–464 (2021).
Kagey, M. H. et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature 467, 430–435 (2010).
Dixon, J. R. et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376–380 (2012).
Guo, Y. et al. CTCF/cohesin-mediated DNA looping is required for protocadherin α promoter choice. Proc. Natl Acad. Sci. USA. 109, 21081–21086 (2012).
Nora, E. P. et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature 485, 381–385 (2012).
Sexton, T. et al. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell 148, 458–472 (2012).
Rao, S. S. P. et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159, 1665–1680 (2014).
Schoenfelder, S. & Fraser, P. Long-range enhancer-promoter contacts in gene expression control. Nat. Rev. Genet. 20, 437–455 (2019).
Thiecke, M. J. et al. Cohesin-dependent and -independent mechanisms mediate chromosomal contacts between promoters and enhancers. Cell Rep. 32, 107929 (2020).
Zhang, Y. et al. The fundamental role of chromatin loop extrusion in physiological V(D)J recombination. Nature 573, 600–604 (2019).
Ba, Z. et al. CTCF orchestrates long-range cohesin-driven V(D)J recombinational scanning. Nature 586, 305–310 (2020).
Hill, L. et al. Wapl repression by Pax5 promotes V gene recombination by Igh loop extrusion. Nature 584, 142–147 (2020).
Peters, J.-M. How DNA loop extrusion mediated by cohesin enables V(D)J recombination. Curr. Opin. Cell Biol. 70, 75–83 (2021).
Naumova, N. et al. Organization of the mitotic chromosome. Science 342, 948–953 (2013).
Gibcus, J. H. et al. A pathway for mitotic chromosome formation. Science 359, eaao6135 (2018).
Batty, P. & Gerlich, D. W. Mitotic chromosome mechanics: how cells segregate their genome. Trends Cell Biol. 29, 717–726 (2019).
Lupiáñez, D. G. et al. Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell 161, 1012–1025 (2015).
Franke, M. et al. Formation of new chromatin domains determines pathogenicity of genomic duplications. Nature 538, 265–269 (2016).
Gröschel, S. et al. A single oncogenic enhancer rearrangement causes concomitant EVI1 and GATA2 deregulation in leukemia. Cell 157, 369–381 (2014).
Northcott, P. A. et al. Enhancer hijacking activates GFI1 family oncogenes in medulloblastoma. Nature 511, 428–434 (2014).
Dekker, J., Rippe, K., Dekker, M. & Kleckner, N. Capturing chromosome conformation. Science 295, 1306–1311 (2002).
McCord, R. P., Kaplan, N. & Giorgetti, L. Chromosome conformation capture and beyond: toward an integrative view of chromosome structure and function. Mol. Cell 77, 688–708 (2020).
Lieberman-Aiden, E. et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326, 289–293 (2009).
Belaghzal, H., Dekker, J. & Gibcus, J. H. Hi-C 2.0: an optimized Hi-C procedure for high-resolution genome-wide mapping of chromosome conformation. Methods 123, 56–65 (2017).
Dekker, J., Marti-Renom, M. A. & Mirny, L. A. Exploring the three-dimensional organization of genomes: interpreting chromatin interaction data. Nat. Rev. Genet. 14, 390–403 (2013).
Yatskevich, S., Rhodes, J. & Nasmyth, K. Organization of chromosomal DNA by SMC complexes. Annu. Rev. Genet. 53, 445–482 (2019).
Hustedt, N. & Durocher, D. The control of DNA repair by the cell cycle. Nat. Cell Biol. 19, 1–9 (2016).
Scully, R., Panday, A., Elango, R. & Willis, N. A. DNA double-strand break repair-pathway choice in somatic mammalian cells. Nat. Rev. Mol. Cell Biol. 20, 698–714 (2019).
Goloborodko, A., Imakaev, M. V., Marko, J. F. & Mirny, L. Compaction and segregation of sister chromatids via active loop extrusion. Elife 5, e14864 (2016).
Mitter, M. et al. Conformation of sister chromatids in the replicated human genome. Nature 586, 139–144 (2020).
Herzog, V. A. et al. Thiol-linked alkylation of RNA to assess expression dynamics. Nat. Methods 14, 1198–1204 (2017).
Riml, C. et al. Osmium-mediated transformation of 4-thiouridine to cytidine as key to study RNA dynamics by sequencing. Angew. Chem. Int. Ed. Engl. 56, 13479–13483 (2017).
Lusser, A. et al. Thiouridine-to-cytidine conversion sequencing (TUC-Seq) to measure mRNA transcription and degradation rates. Methods Mol. Biol. 2062, 191–211 (2020).
Jeppsson, K., Kanno, T., Shirahige, K. & Sjögren, C. The maintenance of chromosome structure: positioning and functioning of SMC complexes. Nat. Rev. Mol. Cell Biol. 15, 601–614 (2014).
Jeppsson, K. et al. The chromosomal association of the Smc5/6 complex depends on cohesion and predicts the level of sister chromatid entanglement. PLoS Genet. 10, e1004680 (2014).
Kadyk, L. C. & Hartwell, L. H. Sister chromatids are preferred over homologs as substrates for recombinational repair in Saccharomyces cerevisiae. Genetics 132, 387–402 (1992).
Liang, F., Han, M., Romanienko, P. J. & Jasin, M. Homology-directed repair is a major double-strand break repair pathway in mammalian cells. Proc. Natl Acad. Sci. USA. 95, 5172–5177 (1998).
Aymard, F. et al. Transcriptionally active chromatin recruits homologous recombination at DNA double-strand breaks. Nat. Struct. Mol. Biol. 21, 366–374 (2014).
Clouaire, T. et al. Comprehensive mapping of histone modifications at DNA double-strand breaks deciphers repair pathway chromatin signatures. Mol. Cell 72, 250–262.e6 (2018).
Schep, R. et al. Impact of chromatin context on Cas9-induced DNA double-strand break repair pathway balance. Mol. Cell 81, 2216–2230.e10 (2021).
Sjögren, C. & Nasmyth, K. Sister chromatid cohesion is required for postreplicative double-strand break repair in Saccharomyces cerevisiae. Curr. Biol. 11, 991–995 (2001).
Potts, P. R., Porteus, M. H. & Yu, H. Human SMC5/6 complex promotes sister chromatid homologous recombination by recruiting the SMC1/3 cohesin complex to double-strand breaks. EMBO J. 25, 3377–3388 (2006).
Watrin, E. & Peters, J.-M. Cohesin and DNA damage repair. Exp. Cell Res. 312, 2687–2693 (2006).
Waizenegger, I. C., Hauf, S., Meinke, A. & Peters, J. M. Two distinct pathways remove mammalian cohesin from chromosome arms in prophase and from centromeres in anaphase. Cell 103, 399–410 (2000).
Losada, A., Hirano, M. & Hirano, T. Cohesin release is required for sister chromatid resolution, but not for condensin-mediated compaction, at the onset of mitosis. Genes Dev. 16, 3004–3016 (2002).
Hirota, T., Gerlich, D., Koch, B., Ellenberg, J. & Peters, J.-M. Distinct functions of condensin I and II in mitotic chromosome assembly. J. Cell Sci. 117, 6435–6445 (2004).
Gerlich, D., Hirota, T., Koch, B., Peters, J.-M. & Ellenberg, J. Condensin I stabilizes chromosomes mechanically through a dynamic interaction in live cells. Curr. Biol. 16, 333–344 (2006).
Zhiteneva, A. et al. Mitotic post-translational modifications of histones promote chromatin compaction in vitro. Open Biol. 7, 170076 (2017).
Ginno, P. A., Burger, L., Seebacher, J., Iesmantavicius, V. & Schübeler, D. Cell cycle-resolved chromatin proteomics reveals the extent of mitotic preservation of the genomic regulatory landscape. Nat. Commun. 9, 4048 (2018).
Zhang, H. et al. Chromatin structure dynamics during the mitosis-to-G1 phase transition. Nature 576, 158–162 (2019).
Abramo, K. et al. A chromosome folding intermediate at the condensin-to-cohesin transition during telophase. Nat. Cell Biol. 21, 1393–1402 (2019).
Kempfer, R. & Pombo, A. Methods for mapping 3D chromosome architecture. Nat. Rev. Genet. 21, 207–226 (2020).
Jerkovic, I. & Cavalli, G. Understanding 3D genome organization by multidisciplinary methods. Nat. Rev. Mol. Cell Biol. 22, 511–528 (2021).
Nagasaka, K., Hossain, M. J., Roberti, M. J., Ellenberg, J. & Hirota, T. Sister chromatid resolution is an intrinsic part of chromosome organization in prophase. Nat. Cell Biol. 18, 692–699 (2016).
Falconer, E. et al. Identification of sister chromatids by DNA template strand sequences. Nature 463, 93–97 (2010).
Chen, B. et al. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell 155, 1479–1491 (2013).
Stanyte, R. et al. Dynamics of sister chromatid resolution during cell cycle progression. J. Cell Biol. 217, 1985–2004 (2018).
Oomen, M. E., Hedger, A. K., Watts, J. K. & Dekker, J. Detecting chromatin interactions between and along sister chromatids with SisterC. Nat. Methods 17, 1002–1009 (2020).
Mitter, M. & Gerlich, D. W. Mapping sister chromatid conformation in replicated chromosomes. Trends Biochem. Sci. 46, 169–170 (2021).
Mumbach, M. R. et al. HiChIP: efficient and sensitive analysis of protein-directed genome architecture. Nat. Methods 13, 919–922 (2016).
Nishimura, K., Fukagawa, T., Takisawa, H., Kakimoto, T. & Kanemaki, M. An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat. Methods 6, 917–922 (2009).
Holland, A. J., Fachinetti, D., Han, J. S. & Cleveland, D. W. Inducible, reversible system for the rapid and complete degradation of proteins in mammalian cells. Proc. Natl Acad. Sci. USA. 109, E3350–E3357 (2012).
Natsume, T., Kiyomitsu, T., Saga, Y. & Kanemaki, M. T. Rapid protein depletion in human cells by auxin-inducible degron tagging with short homology donors. Cell Rep. 15, 210–218 (2016).
Yesbolatova, A. et al. The auxin-inducible degron 2 technology provides sharp degradation control in yeast, mammalian cells, and mice. Nat. Commun. 11, 5701 (2020).
Kurtzer, G. M., Sochat, V. & Bauer, M. W. Singularity: scientific containers for mobility of compute. PLoS One 12, e0177459 (2017).
Di Tommaso, P. et al. Nextflow enables reproducible computational workflows. Nat. Biotechnol. 35, 316–319 (2017).
Abdennur, N. & Mirny, L. A. Cooler: scalable storage for Hi-C data and other genomically labeled arrays. Bioinformatics 36, 311–316 (2020).
Langer, C. C. H. & Mitter, M. Container with Tools to Analyze scsHi-C Data. Available at https://zenodo.org/record/5743325#.YgsVdO7MK8U (2021).
Langer, C. C. H. scsHi-C Preprocessing Nextflow Pipeline (2021); https://zenodo.org/record/5742764#.YhYHEejMJZc
Mitter, M. & Langer, C. C. H. A Collection of NGS Analysis Tools. Available at https://zenodo.org/record/5742702#.YgsWI-7MK8U (2021).
Mitter, M. & Langer, C. C. H. HiglassUp: A higlass Upload Tool. Available at https://zenodo.org/record/5743331#.YgsWee7MK8U (2021).
Held, M. et al. CellCognition: time-resolved phenotype annotation in high-throughput live cell imaging. Nat. Methods 7, 747–754 (2010).
Sommer, C., Hoefler, R., Samwer, M. & Gerlich, D. W. A deep learning and novelty detection framework for rapid phenotyping in high-content screening. Mol. Biol. Cell 28, 3428–3436 (2017).
Hande, K. R. Etoposide: four decades of development of a topoisomerase II inhibitor. Eur. J. Cancer 34, 1514–1521 (1998).
Mitter, M. & Langer, C. C. H. scsHi-C Analysis Notebooks. Available at https://zenodo.org/record/5742704#.YgsW2u7MK8U (2021).
Imakaev, M. et al. Iterative correction of Hi-C data reveals hallmarks of chromosome organization. Nat. Methods 9, 999–1003 (2012).
The HDF Group. Hierarchical Data Format Version 5. Available at http://www.hdfgroup.org/HDF5 (2006).
Kerpedjiev, P. et al. HiGlass: web-based visual exploration and analysis of genome interaction maps. Genome Biol. 19, 125 (2018).
Lajoie, B. R., Dekker, J. & Kaplan, N. The Hitchhiker’s guide to Hi-C analysis: practical guidelines. Methods 72, 65–75 (2015).
Erceg, J. et al. The genome-wide multi-layered architecture of chromosome pairing in early Drosophila embryos. Nat. Commun. 10, 1–13 (2019).
Juicer and Juicebox for chromatin conformation analysis. Nat. Methods 13, 816 (2016).
An, L. et al. OnTAD: hierarchical domain structure reveals the divergence of activity among TADs and boundaries. Genome Biol. 20, 282 (2019).
Yang, T. et al. HiCRep: assessing the reproducibility of Hi-C data using a stratum-adjusted correlation coefficient. Genome Res. 27, 1939–1949 (2017).
Gassler, J. et al. A mechanism of cohesin-dependent loop extrusion organizes zygotic genome architecture. EMBO J. 36, 3600–3618 (2017).
Rao, S. S. P. et al. Cohesin loss eliminates all loop domains. Cell 171, 305–320.e24 (2017).
Acknowledgements
The authors acknowledge technical support by the IMBA/IMP/GMI BioOptics and Molecular Biology Services facilities and the Vienna BioCenter Metabolomics and Next Generation Sequencing facilities. Research in the laboratory of D.W.G. is supported by the Austrian Academy of Sciences, the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (grant agreement No. 101019039), the Austrian Science Fund (FWF; Doktoratskolleg ‘Chromosome Dynamics’ DK W1238) and the Vienna Science and Technology Fund (WWTF; projects LS17-003 and LS19-001). Research in the laboratory of R.M. is supported by the Austrian Science Fund (P31691 and F8011), the Austrian Research Promotion Agency FFG (West-Austrian BioNMR 858017) and the WWTF (project nr. LS17-003). M.M. received a PhD fellowship from the Boehringer Ingelheim Fonds. Z.T. received a Hertha Firnberg Programme fellowship of the FWF (T 1246). The VBCF Metabolomics Facility is funded by the City of Vienna through the Vienna Business Agency.
Author information
Authors and Affiliations
Contributions
M.M. developed the protocol for scsHi-C, with help from R.M. (4sT conversion chemistry), T.K. (mass spectrometry), C.C.H.L. (data processing) and D.W.G. (biological interpretation). M.M., Z.T. and D.W.G. wrote the manuscript, except the procedures section on mass spectrometry, which was written by T.K. D.W.G., M.M., Z.T. and R.M. acquired funding.
Corresponding authors
Ethics declarations
Competing interests
R.M. is listed as inventor on a patent application that has been filed concerning the nucleoside conversion chemistry of this work (Osmiumtetroxide-based conversion of RNA and DNA containing thiolated nucleotides, US Patent App. 16/533,988). The other authors declare no competing interests.
Peer review
Peer review information
Nature Protocols thanks Feng Yue and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key reference using this protocol
Mitter, M. et al. Nature 586, 139–144 (2020): https://doi.org/10.1038/s41586-020-2744-4
Extended data
Extended Data Fig. 1 Yield of scsHi-C using HeLa Kyoto cells and 4sT incorporation in other cell lines.
a, Yield of scsHi-C performed by using HeLa Kyoto cells at different steps of the scsHi-C protocol. b, Quantification of 4sT incorporation into genomic DNA of HCT116, HEK293 and RPE1 cells. Cells were cultured in the presence of 2 mM 4sT for 5 d, and genomic DNA was purified, digested into nucleosides and analyzed by mass spectrometry. The percentage of 4sT over total thymidine is shown from n = 2 biologically independent experiments for each cell line.
Extended Data Fig. 2 HPLC-tandem MS chromatograms of a separated standard mixture of 4sT and thymidine.
Top panels, SRM (selected reaction monitoring) traces of dT (m/z 243.1 to m/z 127.1) and of its in-source fragmentation product (m/z 127 to m/z 54) are shown. Lower panels, SRM traces of thio-dT (m/z 259.1 to m/z 143.1) and the respective in-source fragmentation product (m/z 127.1 to m/z 54.1) are depicted. Both nucleosides generate only one fragment ion (neutral loss of the sugar); therefore, we recommend recording the SRM traces of the in-source products, which can either be used for quantification (if of significant signal intensity) or as a qualifier for confirming the respective nucleoside.
Rights and permissions
About this article
Cite this article
Mitter, M., Takacs, Z., Köcher, T. et al. Sister chromatid–sensitive Hi-C to map the conformation of replicated genomes. Nat Protoc 17, 1486–1517 (2022). https://doi.org/10.1038/s41596-022-00687-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-022-00687-6
This article is cited by
-
Time-resolved single-cell RNA-seq using metabolic RNA labelling
Nature Reviews Methods Primers (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.