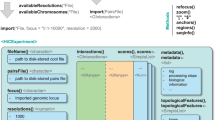
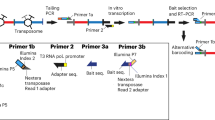
Abstract
Chromosome conformation capture (3C) methods measure the spatial proximity between DNA elements in the cell nucleus. Many methods have been developed to sample 3C material, including the Capture-C family of protocols. Capture-C methods use oligonucleotides to enrich for interactions of interest from sequencing-ready 3C libraries. This approach is modular and has been adapted and optimized to work for sampling of disperse DNA elements (NuTi Capture-C), including from low cell inputs (LI Capture-C), as well as to generate Hi-C like maps for specific regions of interest (Tiled-C) and to interrogate multiway interactions (Tri-C). We present the design, experimental protocol and analysis pipeline for NuTi Capture-C in addition to the variations for generation of LI Capture-C, Tiled-C and Tri-C data. The entire procedure can be performed in 3 weeks and requires standard molecular biology skills and equipment, access to a next-generation sequencing platform, and basic bioinformatic skills. Implemented with other sequencing technologies, these methods can be used to identify regulatory interactions and to compare the structural organization of the genome in different cell types and genetic models.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Code availability
CapCruncher can be used following direct installation from Bioconda or accessed via GitHub91 (https://github.com/sims-lab/CapCruncher/releases).
Change history
27 June 2023
A Correction to this paper has been published: https://doi.org/10.1038/s41596-023-00860-5
References
Dekker, J., Rippe, K., Dekker, M. & Kleckner, N. Capturing chromosome conformation. Science 295, 1306–1311 (2002).
Brant, L. et al. Exploiting native forces to capture chromosome conformation in mammalian cell nuclei. Mol. Syst. Biol. 12, 1–8 (2016).
Rao, S. S. P. et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159, 1665–1680 (2014).
Hughes, J. R. et al. Analysis of hundreds of cis-regulatory landscapes at high resolution in a single, high-throughput experiment. Nat. Genet. 46, 205–212 (2014).
Davies, J. O. J. et al. Multiplexed analysis of chromosome conformation at vastly improved sensitivity. Nat. Methods 13, 74–80 (2016).
Van De Werken, H. J. G. et al. Robust 4C-seq data analysis to screen for regulatory DNA interactions. Nat. Methods 9, 969–972 (2012).
Mifsud, B. et al. Mapping long-range promoter contacts in human cells with high-resolution capture Hi-C. Nat. Genet. 47, 598–606 (2015).
Madsen, J. G. S. et al. Highly interconnected enhancer communities control lineage-determining genes in human mesenchymal stem cells. Nat. Genet. 52, 1227–1238 (2020).
Oudelaar, A. M., Davies, J. O. J., Downes, D. J., Higgs, D. R. & Hughes, J. R. Robust detection of chromosomal interactions from small numbers of cells using low-input Capture-C. Nucleic Acids Res. 45, (2017).
Downes, D. J. et al. High-resolution targeted 3C interrogation of cis-regulatory element organization at genome-wide scale. Nat. Commun. 12, 531 (2021).
Oudelaar, A. M. et al. Single-allele chromatin interactions identify regulatory hubs in dynamic compartmentalized domains. Nat. Genet. 50, 1744–1751 (2018).
Oudelaar, A. M., Hughes, J. & Downes, D. Tri-C. Protoc. Exch. https://doi.org/10.21203/rs.2.1650/v2 (2019).
Oudelaar, A. M. et al. Dynamics of the 4D genome during in vivo lineage specification and differentiation. Nat. Commun. 11, (2020).
Golov, A. K. et al. A modified protocol of Capture-C allows affordable and flexible high-resolution promoter interactome analysis. Sci. Rep. 10, 1–15 (2020).
King, A. J. et al. Reactivation of a developmentally silenced embryonic globin gene. Nat. Commun. https://doi.org/10.1038/s41467-021-24402-3 (2021).
Hay, D. et al. Genetic dissection of the α-globin super-enhancer in vivo. Nat. Genet. 48, 895–903 (2016).
Simon, C. S. et al. Functional characterisation of cis-regulatory elements governing dynamic Eomes expression in the early mouse embryo. Development 144, 1249–1260 (2017).
Schäfer, A. et al. Impaired DNA demethylation of C/EBP sites causes premature aging. Genes Dev. 32, 742–762 (2018).
Godfrey, L. et al. DOT1L inhibition reveals a distinct subset of enhancers dependent on H3K79 methylation. Nat. Commun. 10, 2803 (2019).
Oudelaar, A. M. et al. A revised model for promoter competition based on multi-way chromatin interactions at the α-globin locus. Nat. Commun. https://doi.org/10.1038/s41467-019-13404-x (2019).
Ghavi-Helm, Y. et al. Highly rearranged chromosomes reveal uncoupling between genome topology and gene expression. Nat. Genet. 51, 1272–1282 (2019).
Williams, R. M. et al. Reconstruction of the global neural crest gene regulatory network in vivo. Dev. Cell 51, 255–276.e7 (2019).
Larke, M. S. C. et al. Enhancers predominantly regulate gene expression during differentiation via transcription initiation. Mol. Cell 81, 983-997.e7 (2021).
Blackledge, N. P. et al. PRC1 catalytic activity is central to polycomb system function. Mol. Cell 77, 857-874.e9 (2020).
Rhodes, J. D. P. et al. Cohesin disrupts polycomb-dependent chromosome interactions in embryonic stem cells. Cell Rep. 30, 820–835 (2020).
Furlan, G. et al. The Ftx noncoding locus controls X chromosome inactivation independently of its RNA products. Mol. Cell 70, 462–472 (2018).
van Bemmel, J. G. et al. The bipartite TAD organization of the X-inactivation center ensures opposing developmental regulation of Tsix and Xist. Nat. Genet. 51, 1024–1034 (2019).
Hanssen, L. L. P. et al. Tissue-specific CTCF–cohesin-mediated chromatin architecture delimits enhancer interactions and function in vivo. Nat. Cell Biol. 19, 952–961 (2017).
Hyle, J. et al. Acute depletion of CTCF directly affects MYC regulation through loss of enhancer–promoter looping. Nucleic Acids Res. 47, 6699–6713 (2019).
Zhang, D. et al. Alteration of genome folding via contact domain boundary insertion. Nat. Genet. 52, 1076-1087 (2020).
Harrold, C. L. et al. A functional overlap between actively transcribed genes and chromatin boundary elements. Preprint at bioRxiv https://doi.org/10.1101/2020.07.01.182089 (2020).
Downes, D. J. et al. An integrated platform to systematically identify causal variants and genes for polygenic human traits. Preprint at bioRxiv https://doi.org/10.1101/813618 (2019).
Thurner, M. et al. Integration of human pancreatic islet genomic data refines regulatory mechanisms at Type 2 diabetes susceptibility loci. eLife 7, e31977 (2018).
Chesi, A. et al. Genome-scale Capture C promoter interactions implicate effector genes at GWAS loci for bone mineral density. Nat. Commun. 10, 1260 (2019).
Badat, M. et al. A remarkable case of HbH disease illustrates the relative contributions of the α-globin enhancers to gene expression. Blood https://doi.org/10.1182/blood.2020006680 (2020).
Long, H. K. et al. Loss of extreme long-range enhancers in human neural crest drives a craniofacial disorder. Cell Stem Cell 27, 765–783.e14 (2020).
Olijnik, A. A. et al. Genetic and functional insights into CDA-I prevalence and pathogenesis. J. Med. Genet. https://doi.org/10.1136/jmedgenet-2020-106880 (2020).
Bozhilov, Y. K. et al. A gain-of-function single nucleotide variant creates a new promoter which acts as an orientation-dependent enhancer–blocker. Nat. Commun. 12, 3806 (2021).
Schwessinger, R. et al. DeepC: predicting 3D genome folding using megabase-scale transfer learning. Nat. Methods https://doi.org/10.1038/s41592-020-0960-3 (2020).
Brown, J. M. et al. A tissue-specific self-interacting chromatin domain forms independently of enhancer–promoter interactions. Nat. Commun. 9, 3849 (2018).
Chiariello, A. M. et al. A dynamic folded hairpin conformation is associated with α-globin activation in erythroid cells. Cell Rep. 30, 2125–2135.e5 (2020).
Zhao, Z. et al. Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nat. Genet. 38, 1341–1347 (2006).
Simonis, M. et al. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture–on-chip (4C). Nat. Genet. 38, 1348–1354 (2006).
Hagege, H. et al. Quantitative analysis of chromosome conformation capture assays (3C-qPCR). Nat. Protoc. 2, 1722–1733 (2007).
Schwartzman, O. et al. UMI-4C for quantitative and targeted chromosomal contact profiling. Nat. Methods 13, 685–691 (2016).
Davies, J. O. J., Oudelaar, A. M., Higgs, D. R. & Hughes, J. R. How best to identify chromosomal interactions: a comparison of approaches. Nat. Methods 14, 125–134 (2017).
Schoenfelder, S. et al. The pluripotent regulatory circuitry connecting promoters to their long-range interacting elements. Genome Res. 25, 582–597 (2015).
Hsieh, T. H. S. et al. Mapping nucleosome resolution chromosome folding in yeast by Micro-C. Cell 162, 108–119 (2015).
Hsieh, T. H. S., Fudenberg, G., Goloborodko, A. & Rando, O. J. Micro-C XL: assaying chromosome conformation from the nucleosome to the entire genome. Nat. Methods 13, 1009–1011 (2016).
Ma, W. et al. Fine-scale chromatin interaction maps reveal the cis-regulatory landscape of human lincRNA genes. Nat. Methods 12, 71–78 (2014).
Hua, P. et al. Defining genome architecture at base-pair resolution. Nature https://doi.org/10.1038/s41586-021-03639-4 (2021).
Li, G. et al. Chromatin interaction analysis with paired-end tag (ChIA-PET) sequencing technology and application. BMC Genomics 15, S11 (2014).
Fang, R. et al. Mapping of long-range chromatin interactions by proximity ligation-assisted ChIP-seq. Cell Res. 26, 1345–1348 (2016).
Zheng, M. et al. Multiplex chromatin interactions with single-molecule precision. Nature 566, 558–562 (2019).
Mumbach, M. R. et al. HiChIP: efficient and sensitive analysis of protein-directed genome architecture. Nat. Methods 13, 919–922 (2016).
Mumbach, M. R. et al. HiChIRP reveals RNA-associated chromosome conformation. Nat. Methods 16, 489–492 (2019).
Dostie, J. et al. Chromosome Conformation Capture Carbon Copy (5C): a massively parallel solution for mapping interactions between genomic elements. Genome Res. 16, 1299–1309 (2006).
Kolovos, P. et al. Targeted chromatin capture (T2C): a novel high resolution high throughput method to detect genomic interactions and regulatory elements. Epigenetics Chromatin 7, 10 (2014).
Dryden, N. H. et al. Unbiased analysis of potential targets of breast cancer susceptibility loci by Capture Hi-C. Genome Res. 24, 1854–1868 (2014).
Sanborn, A. L. et al. Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proc. Natl Acad. Sci. USA 112, E6456–E6465 (2015).
Aljahani, A. et al. Analysis of sub-kilobase chromatin topology reveals nano-scale regulatory interactions with variable dependence on cohesin and CTCF. Preprint at bioRxiv https://doi.org/10.1101/2021.08.10.455796 (2021).
Olivares-Chauvet, P. et al. Capturing pairwise and multi-way chromosomal conformations using chromosomal walks. Nature 540, 296–300 (2016).
Allahyar, A. et al. Enhancer hubs and loop collisions identified from single-allele topologies. Nat. Genet. 50, 1151–1160 (2018).
Vermeulen, C. et al. Multi-contact 4C: long-molecule sequencing of complex proximity ligation products to uncover local cooperative and competitive chromatin topologies. Nat. Protoc. 15, 364–397 (2020).
Beagrie, R. A. et al. Multiplex-GAM: genome-wide identification of chromatin contacts yields insights not captured by Hi-C. Preprint at bioRxiv https://doi.org/10.1101/2020.07.31.230284 (2020).
Beagrie, R. A. et al. Complex multi-enhancer contacts captured by genome architecture mapping. Nature 543, 519–524 (2017).
Quinodoz, S. A. et al. Higher-order inter-chromosomal hubs shape 3D genome organization in the nucleus. Cell 174, 744–757.e24 (2018).
Takei, Y. et al. Integrated spatial genomics reveals global architecture of single nuclei. Nature 590, 344–350 (2021).
Tan, L., Xing, D., Chang, C. H., Li, H. & Xie, X. S. Three-dimensional genome structures of single diploid human cells. Science 361, 924–928 (2018).
Nagano, T. et al. Single-cell Hi-C reveals cell-to-cell variability in chromosome structure. Nature 502, 59–64 (2013).
Nagano, T. et al. Cell-cycle dynamics of chromosomal organization at single-cell resolution. Nature 547, 61–67 (2017).
Ramani, V. et al. Massively multiplex single-cell Hi-C. Nat. Methods 14, 263–266 (2017).
Stevens, T. J. et al. 3D structures of individual mammalian genomes studied by single-cell Hi-C. Nature 544, 59–64 (2017).
Flyamer, I. M. et al. Single-nucleus Hi-C reveals unique chromatin reorganization at oocyte-to-zygote transition. Nature 544, 110–114 (2017).
Telenius, J. M. et al. CaptureCompendium: a comprehensive toolkit for 3C analysis. Preprint at bioRxiv https://doi.org/10.1101/2020.02.17.952572 (2020).
Anil, A., Spalinskas, R., Åkerborg, Ö. & Sahlén, P. HiCapTools: a software suite for probe design and proximity detection for targeted chromosome conformation capture applications. Bioinformatics 34, 675–677 (2018).
Hansen, P. et al. GOPHER: Generator Of probes for capture Hi-C experiments at high resolution. BMC Genomics 20, 40 (2019).
Kent, W. J. BLAT—the BLAST-like alignment tool. Genome Res. 12, 656–664 (2002).
Smit, A., Hubley, R. & Green, P. RepeatMasker Open-4.0 (2015).
Eijsbouts, C. Q., Burren, O. S., Newcombe, P. J. & Wallace, C. Fine mapping chromatin contacts in capture Hi-C data. BMC Genomics 20, 77 (2019).
Geeven, G., Teunissen, H., De Laat, W. & De Wit, E. peakC: a flexible, non-parametric peak calling package for 4C and Capture-C data. Nucleic Acids Res. 46, e91 (2018).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 1–21 (2014).
Wang, Y. et al. The 3D Genome Browser: a web-based browser for visualizing 3D genome organization and long-range chromatin interactions. Genome Biol. https://doi.org/10.1101/112268 (2018).
Kerpedjiev, P. et al. HiGlass: web-based visual exploration and analysis of genome interaction maps. Genome Biol. https://doi.org/10.1101/121889 (2018).
Servant, N. et al. HiC-Pro: an optimized and flexible pipeline for Hi-C data processing. Genome https://doi.org/10.1186/s13059-015-0831-x (2015).
Buckle, A., Gilbert, N., Marenduzzo, D. & Brackley, C. A. capC-MAP: software for analysis of Capture-C data. Bioinformatics https://doi.org/10.1093/bioinformatics/btz480 (2019).
Cairns, J. et al. CHiCAGO: robust detection of DNA looping interactions in Capture Hi-C data. Genome Biol. 17, 127 (2016).
Thongjuea, S., Stadhouders, R., Grosveld, F. G., Soler, E. & Lenhard, B. R3Cseq: an R/Bioconductor package for the discovery of long-range genomic interactions from chromosome conformation capture and next-generation sequencing data. Nucleic Acids Res. 41, e132 (2013).
Klein, F. A. et al. FourCSeq: analysis of 4C sequencing data. Bioinformatics 31, 3085–3091 (2015).
Freire-Pritchett, P. et al. Detecting chromosomal interactions in Capture Hi-C data with CHiCAGO and companion tools. Nat. Protoc. 16, 4144–4176 (2021).
Smith, A. L., Rue-Albrecht, K. & Sims, D. CapCruncher. Zenodo https://doi.org/10.5281/zenodo.5113088 (2021).
Brandão, H. B., Gabriele, M. & Hansen, A. S. Tracking and interpreting long-range chromatin interactions with super-resolution live-cell imaging. Curr. Opin. Cell Biol. 70, 18–26 (2021).
Lakadamyali, M. & Cosma, M. P. Visualizing the genome in high resolution challenges our textbook understanding. Nat. Methods 17, 371–379 (2020).
Kempfer, R. & Pombo, A. Methods for mapping 3D chromosome architecture. Nat. Rev. Genet. 21, 207–226 (2020).
Shaban, H. A. & Seeber, A. Monitoring the spatio-temporal organization and dynamics of the genome. Nucleic Acids Res. 48, 3423–3434 (2020).
Beecham, A. H. et al. Analysis of immune-related loci identifies 48 new susceptibility variants for multiple sclerosis. Nat. Genet. 45, 1353–1360 (2013).
Boettiger, A. & Murphy, S. Advances in chromatin imaging at kilobase-scale resolution. Trends Genet 36, 273–287 (2020).
Buenrostro, J. D., Giresi, P. G., Zaba, L. C., Chang, H. Y. & Greenleaf, W. J. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218 (2013).
Boyle, A. P. et al. High-resolution mapping and characterization of open chromatin across the genome. Cell 132, 311–322 (2008).
Oudelaar, A. M., Downes, D., Davies, J. & Hughes, J. Low-input Capture-C: a chromosome conformation capture assay to analyze chromatin architecture in small numbers of cells. Bio Protoc. 7, e2645 (2017).
Acknowledgements
We thank all of our collaborators who provided feedback and sought guidance when using this protocol. These methods were developed as part of the Wellcome Investigation of Genome Wide Association Mechanisms (WIGWAM) Consortium funded by a Wellcome Strategic Award (106130/Z/14/Z). J.R.H. received Medical Research Council (MRC) Core Funding (MC_UU_00016/14). T.A.M. and A.L.S. are supported by Molecular Haematology Unit grant MC_UU_00016/6. J.O.J.D. was supported by grants from Wellcome (098931/Z/12/Z) and the MRC (MR/R008108/1). D.S. received Wellcome funding (204826/Z/16/Z). A.M.O. is supported by the Max Planck Society. M.A.K. and T.V. are supported by the International Max Planck Research School for Genome Science, part of the Göttingen Graduate Center for Neurosciences, Biophysics and Molecular Biosciences.
Author information
Authors and Affiliations
Contributions
J.O.J.D. and J.R.H. designed the original protocol. D.J.D., M.A.K., T.V. and A.M.O. performed optimization experiments and developed the protocol. D.J.D., A.L.S., J.O.J.D., K.R., D.S. and A.M.O. designed and created the data analysis scripts. D.S., T.A.M., A.M.O. and J.R.H. acquired funding and oversaw the work. D.J.D. and A.M.O. wrote the manuscript and generated the figures. All authors critically evaluated and edited the manuscript.
Corresponding authors
Ethics declarations
Competing interests
J.R.H. and J.O.J.D. are founders and shareholders of Nucleome Therapeutics. J.R.H., J.O.J.D. and D.J.D. are paid consultants for Nucleome Therapeutics. J.R.H. and J.O.J.D. hold patents for Capture-C (WO2017068379A1, EP3365464B1, US10934578B2) and have a patent application for MCC. T.A.M. is a founding shareholder of OxStem Oncology (a subsidiary company of OxStem Ltd.) and a founding shareholder and paid consultant for Sandymount Therapeutics (a subsidiary company of Dark Blue Therapeutics). The other authors have no competing interests.
Peer review
Peer review information
Nature Protocols thanks Zhijun Duan and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key references using this protocol
Downes, D. J. et al. Nat. Commun. 12, 531 (2021): https://doi.org/10.1038/s41467-020-20809-6
Oudelaar, A. M. et al. Nat. Commun. 11, 2722 (2020): https://doi.org/10.1038/s41467-020-16598-7
Oudelaar, A. M. et al. Nat. Genet. 50, 1744–1751 (2018): https://doi.org/10.1038/s41588-018-0253-2
Supplementary information
Supplementary Information
Supplementary Manual
Supplementary Data 1
Files for designing NuTi Capture-C viewpoints, analyzing NuTi Capture-C viewpoints and running CapCruncher.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Downes, D.J., Smith, A.L., Karpinska, M.A. et al. Capture-C: a modular and flexible approach for high-resolution chromosome conformation capture. Nat Protoc 17, 445–475 (2022). https://doi.org/10.1038/s41596-021-00651-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-021-00651-w
This article is cited by
-
β-actin mediated H3K27ac changes demonstrate the link between compartment switching and enhancer-dependent transcriptional regulation
Genome Biology (2023)
-
Determining chromatin architecture with Micro Capture-C
Nature Protocols (2023)
-
The Mediator complex regulates enhancer-promoter interactions
Nature Structural & Molecular Biology (2023)
-
MLL-AF4 cooperates with PAF1 and FACT to drive high-density enhancer interactions in leukemia
Nature Communications (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.