Abstract
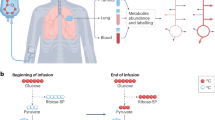
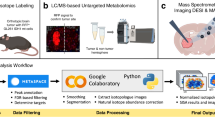
Cancer cells undergo diverse metabolic adaptations to meet the energetic demands imposed by dysregulated growth and proliferation. Assessing metabolism in intact tumors allows the investigator to observe the combined metabolic effects of numerous cancer cell-intrinsic and -extrinsic factors that cannot be fully captured in culture models. We have developed methods to use stable isotope-labeled nutrients (e.g., [13C]glucose) to probe metabolic activity within intact tumors in vivo, in mice and humans. In these methods, the labeled nutrient is introduced to the circulation through an intravenous catheter prior to surgical resection of the tumor and adjacent nonmalignant tissue. Metabolism within these tissues during the infusion transfers the isotope label into metabolic intermediates from pathways supplied by the infused nutrient. Extracting metabolites from surgical specimens and analyzing their isotope labeling patterns provides information about metabolism in the tissue. We provide detailed information about this technique, from introduction of the labeled tracer through data analysis and interpretation, including streamlined approaches to quantify isotope labeling in informative metabolites extracted from tissue samples. We focus on infusions with [13C]glucose and the application of mass spectrometry to assess isotope labeling in intermediates from central metabolic pathways, including glycolysis, the tricarboxylic acid cycle and nonessential amino acid synthesis. We outline practical considerations to apply these methods to human subjects undergoing surgical resections of solid tumors. We also discuss the method’s versatility and consider the relative advantages and limitations of alternative approaches to introduce the tracer, harvest the tissue and analyze the data.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
Previously unpublished data are included in the Supplementary Information. The remaining supporting data can be found in refs. 4,5,21.
Code availability
The software can be found at https://github.com/wencgu/nac.
References
Faubert, B. & DeBerardinis, R. J. Analyzing tumor metabolism in vivo. Annu. Rev. Cancer Biol. 1, 99–117 (2016).
Buescher, J. M. et al. A roadmap for interpreting (13)C metabolite labeling patterns from cells. Curr. Opin. Biotechnol. 34, 189–201 (2015).
Maher, E. A. et al. Metabolism of [U-13 C]glucose in human brain tumors in vivo. NMR Biomed. 25, 1234–1244 (2012).
Hensley, C. T. et al. Metabolic heterogeneity in human lung tumors. Cell 164, 681–694 (2016).
Faubert, B. et al. Lactate metabolism in human lung tumors. Cell 171, 358–371 e359 (2017).
Courtney, K. D. et al. Isotope tracing of human clear cell renal cell carcinomas demonstrates suppressed glucose oxidation in vivo. Cell Metab. 28, 793–800 (2018).
Johnston, K. et al. Isotope tracing reveals glycolysis and oxidative metabolism in childhood tumors of multiple histologies. Med 2, 395–410 (2021).
Oizel, K. et al. Glutamine uptake and utilization of human mesenchymal glioblastoma in orthotopic mouse model. Cancer Metab. 8, 9 (2020).
Momcilovic, M. et al. The GSK3 signaling axis regulates adaptive glutamine metabolism in lung squamous cell carcinoma. Cancer Cell 33, 905–921 e905 (2018).
Davidson, S. M. et al. Environment impacts the metabolic dependencies of Ras-driven non-small cell lung cancer. Cell Metab. 23, 517–528 (2016).
Shi, X. et al. The abundance of metabolites related to protein methylation correlates with the metastatic capacity of human melanoma xenografts. Sci. Adv. 3, eaao5268 (2017).
Grinde, M. T. et al. Glutamine to proline conversion is associated with response to glutaminase inhibition in breast cancer. Breast Cancer Res. 21, 61 (2019).
Fan, T. W. et al. Altered regulation of metabolic pathways in human lung cancer discerned by (13)C stable isotope-resolved metabolomics (SIRM). Mol. Cancer 8, 41 (2009).
Sellers, K. et al. Pyruvate carboxylase is critical for non-small-cell lung cancer proliferation. J Clin. Invest. 125, 687–698 (2015).
Sun, R. C. et al. Noninvasive liquid diet delivery of stable isotopes into mouse models for deep metabolic network tracing. Nat. Commun. 8, 1646 (2017).
Jang, C., Chen, L. & Rabinowitz, J. D. Metabolomics and isotope tracing. Cell 173, 822–837 (2018).
Roussel, R., Carlier, P. G., Robert, J. J., Velho, G. & Bloch, G. 13C/31P NMR studies of glucose transport in human skeletal muscle. Proc. Natl Acad. Sci. USA 95, 1313–1318 (1998).
Romijn, J. A., Coyle, E. F., Sidossis, L. S., Rosenblatt, J. & Wolfe, R. R. Substrate metabolism during different exercise intensities in endurance-trained women. J. Appl. Physiol. 88, 1707–1714 (2000).
Coggan, A. R., Kohrt, W. M., Spina, R. J., Bier, D. M. & Holloszy, J. O. Endurance training decreases plasma glucose turnover and oxidation during moderate-intensity exercise in men. J. Appl. Physiol. 68, 990–996 (1990).
Ma, E. H. et al. Metabolic profiling using stable isotope tracing reveals distinct patterns of glucose utilization by physiologically activated CD8(+) T cells. Immunity 51, 856–870 e855 (2019).
Tasdogan, A. et al. Metabolic heterogeneity confers differences in melanoma metastatic potential. Nature 577, 115–120 (2020).
Marin-Valencia, I. et al. Analysis of tumor metabolism reveals mitochondrial glucose oxidation in genetically diverse human glioblastomas in the mouse brain in vivo. Cell Metab. 15, 827–837 (2012).
Hui, S. et al. Glucose feeds the TCA cycle via circulating lactate. Nature 551, 115–118 (2017).
Pan, M. et al. Regional glutamine deficiency in tumours promotes dedifferentiation through inhibition of histone demethylation. Nat. Cell Biol. 18, 1090–1101 (2016).
Zhang, J. et al. 13C isotope-assisted methods for quantifying glutamine metabolism in cancer cells. Methods Enzymol. 542, 369–389 (2014).
Long, C. P. & Antoniewicz, M. R. High-resolution (13)C metabolic flux analysis. Nat. Protoc. 14, 2856–2877 (2019).
Yuan, M. et al. Ex vivo and in vivo stable isotope labelling of central carbon metabolism and related pathways with analysis by LC-MS/MS. Nat. Protoc. 14, 313–330 (2019).
Broekaert, D. & Fendt, S. M. Measuring in vivo tissue metabolism using (13)C glucose infusions in mice. Methods Mol. Biol. 1862, 67–82 (2019).
Su, X., Lu, W. & Rabinowitz, J. D. Metabolite spectral accuracy on orbitraps. Anal. Chem. 89, 5940–5948 (2017).
Antoniewicz, M. R., Kelleher, J. K. & Stephanopoulos, G. Elementary metabolite units (EMU): a novel framework for modeling isotopic distributions. Metab. Eng. 9, 68–86 (2007).
Millard, P., Letisse, F., Sokol, S. & Portais, J. C. IsoCor: correcting MS data in isotope labeling experiments. Bioinformatics 28, 1294–1296 (2012).
Young, J. D. INCA: a computational platform for isotopically non-stationary metabolic flux analysis. Bioinformatics 30, 1333–1335 (2014).
Alger, J. R., Sherry, A. D. & Malloy, C. R. tcaSIM: a simulation program for optimal design of (13)C tracer experiments for analysis of metabolic flux by NMR and mass spectroscopy. Curr. Metabolomics 6, 176–187 (2018).
Yoo, H., Antoniewicz, M. R., Stephanopoulos, G. & Kelleher, J. K. Quantifying reductive carboxylation flux of glutamine to lipid in a brown adipocyte cell line. J. Biol. Chem. 283, 20621–20627 (2008).
Weitzel, M. et al. 13CFLUX2-high-performance software suite for (13)C-metabolic flux analysis. Bioinformatics 29, 143–145 (2013).
Hui, S. et al. Quantitative fluxomics of circulating metabolites. Cell Metab. 32, 676–688 e674 (2020).
Jang, C. et al. Metabolite exchange between mammalian organs quantified in pigs. Cell Metab. 30, 594–606 e593 (2019).
Liu, S., Dai, Z., Cooper, D. E., Kirsch, D. G. & Locasale, J. W. Quantitative analysis of the physiological contributions of glucose to the TCA cycle. Cell Metab. 32, 619–628 e621 (2020).
Acknowledgements
R.J.D. is a Howard Hughes Medical Institute (HHMI) Investigator, the Robert L. Moody, Sr. Faculty Scholar at UT Southwestern and Joel B. Steinberg, M.D. Distinguished Chair in Pediatrics. S.J.M. is an HHMI Investigator, the Mary McDermott Cook Chair in Pediatric Genetics, the Kathryn and Gene Bishop Distinguished Chair in Pediatric Research, the director of the Hamon Laboratory for Stem Cells and Cancer, and a Cancer Prevention and Research Institute of Texas Scholar. The research was supported by the Cancer Prevention and Research Institute of Texas (RP170114 and RP180778), the National Institutes of Health (R35 CA220449; U01 CA228608) and the Robert A. Welch Foundation (I-1733). A.T. was supported by the Leopoldina Fellowship (LPDS 2016-16) from the German National Academy of Sciences and the Fritz Thyssen Foundation. B.F. is supported by the National Institutes of Health (K99/R00 CA237724-01A1). Figures were generated using BioRender.com.
Author information
Authors and Affiliations
Contributions
B.F., A.T., S.J.M., T.P.M and R.J.D. wrote and edited the manuscript. T.P.M established HRMS methodology.
Corresponding authors
Ethics declarations
Competing interests
R.J.D. is an adviser for Agios Pharmaceuticals and Vida Ventures, and a founder and adviser for Atavistik Bio. S.J.M. is an adviser for Frequency Therapeutics and Protein Fluidics.
Additional information
Peer review information Nature Protocols thanks Monica Montopoli and the other, anonymous reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key data used in this protocol
Faubert, B. et al. Cell 171, 358–371 (2017): https://doi.org/10.1016/j.cell.2017.09.019
Tasdogan, A. et al. Nature 577, 115–120 (2020): https://doi.org/10.1038/s41586-019-1847-2
Hensley, CT. et al. Cell 164, 681–694 (2016): https://doi.org/10.1016/j.cell.2015.12.034
Supplementary information
Supplementary Data 1
Supplementary data tables for patient and mouse 13C glucose infusions
Rights and permissions
About this article
Cite this article
Faubert, B., Tasdogan, A., Morrison, S.J. et al. Stable isotope tracing to assess tumor metabolism in vivo. Nat Protoc 16, 5123–5145 (2021). https://doi.org/10.1038/s41596-021-00605-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-021-00605-2
This article is cited by
-
A Novel Strategy for the Development of Functional Foods to Improve Energy Metabolism Disorders: Stable Isotope-Resolved Metabolomics
Food and Bioprocess Technology (2024)
-
Characterizing cancer metabolism from bulk and single-cell RNA-seq data using METAFlux
Nature Communications (2023)
-
MIRTH: Metabolite Imputation via Rank-Transformation and Harmonization
Genome Biology (2022)
-
Metabolic flux between organs measured by arteriovenous metabolite gradients
Experimental & Molecular Medicine (2022)
-
Metabolic regulation of somatic stem cells in vivo
Nature Reviews Molecular Cell Biology (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.