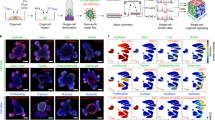
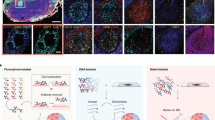
Abstract
Organoids are biomimetic tissue models comprising multiple cell types and cell states. Post-translational modification (PTM) signaling networks control cellular phenotypes and are frequently dysregulated in diseases such as cancer. Although signaling networks vary across cell types, there are limited techniques to study cell type–specific PTMs in heterocellular organoids. Here, we present a multiplexed mass cytometry (MC) protocol for single-cell analysis of PTM signaling and cell states in organoids and organoids co-cultured with fibroblasts and leukocytes. We describe how thiol-reactive organoid barcoding in situ (TOBis) enables 35-plex and 126-plex single-cell comparison of organoid cultures and provide a cytometry by time of flight (CyTOF) signaling analysis pipeline (CyGNAL) for computing cell type–specific PTM signaling networks. The TOBis MC protocol takes ~3 d from organoid fixation to data acquisition and can generate single-cell data for >40 antibodies from millions of cells across 126 organoid cultures in a single MC run.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
All raw data, processed data and working illustrations are available as a Community Cytobank project (https://community.cytobank.org/cytobank/experiments#project-id=1334).
Code availability
The latest CyGNAL pipeline is available at https://github.com/TAPE-Lab/CyGNAL. CyGNAL version 0.2.1 as described in this publication can be found at https://github.com/TAPE-Lab/CyGNAL/releases/tag/v0.2.1. The OT-2 barcode preparation code is available at https://github.com/TAPE-Lab/OT-2-Automated-Barcode-Pipetting. The code in this paper has been peer reviewed.
References
Clevers, H. Modeling development and disease with organoids. Cell 165, 1586–1597 (2016).
Tuveson, D. & Clevers, H. Cancer modeling meets human organoid technology. Science 364, 952–955 (2019).
Sato, T. et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459, 262–265 (2009).
Huch, M. et al. In vitro expansion of single Lgr5+ liver stem cells induced by Wnt-driven regeneration. Nature 494, 247–250 (2013).
Lancaster, M. A. et al. Cerebral organoids model human brain development and microcephaly. Nature 501, 373–379 (2013).
Huch, M. et al. Unlimited in vitro expansion of adult bi-potent pancreas progenitors through the Lgr5/R-spondin axis. EMBO J. 32, 2708–2721 (2013).
van de Wetering, M. et al. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell 161, 933–945 (2015).
Drost, J. & Clevers, H. Organoids in cancer research. Nat. Rev. Cancer 18, 407–418 (2018).
Vlachogiannis, G. et al. Patient-derived organoids model treatment response of metastatic gastrointestinal cancers. Science 359, 920–926 (2018).
Gehart, H. & Clevers, H. Tales from the crypt: new insights into intestinal stem cells. Nat. Rev. Gastroenterol. Hepatol. 16, 19–34 (2019).
Pawson, T. & Scott, J. D. Protein phosphorylation in signaling—50 years and counting. Trends Biochem. Sci. 30, 286–290 (2005).
Deribe, Y. L., Pawson, T. & Dikic, I. Post-translational modifications in signal integration. Nat. Struct. Mol. Biol. 17, 666–672 (2010).
Zhang, J., Yang, P. L. & Gray, N. S. Targeting cancer with small molecule kinase inhibitors. Nat. Rev. Cancer 9, 28–39 (2009).
Miller-Jensen, K., Janes, K. A., Brugge, J. S. & Lauffenburger, D. A. Common effector processing mediates cell-specific responses to stimuli. Nature 448, 604–608 (2007).
Rapsomaniki, M. A. et al. CellCycleTRACER accounts for cell cycle and volume in mass cytometry data. Nat. Commun. 9, 632 (2018).
Qin, X. & Tape, C. J. Deciphering organoids: high-dimensional analysis of biomimetic cultures. Trends Biotechnol. 39, 774–787 (2020).
Qin, X. et al. Cell-type-specific signaling networks in heterocellular organoids. Nat. Methods 17, 335–342 (2020).
Spitzer, M. H. & Nolan, G. P. Mass cytometry: single cells, many features. Cell 165, 780–791 (2016).
Lin, J. R., Fallahi-Sichani, M. & Sorger, P. K. Highly multiplexed imaging of single cells using a high-throughput cyclic immunofluorescence method. Nat. Commun. 6, 8390 (2015).
Tape, C. J. Systems biology analysis of heterocellular signaling. Trends Biotechnol. 34, 627–637 (2016).
Bandura, D. R. et al. Mass cytometry: technique for real time single cell multitarget immunoassay based on inductively coupled plasma time-of-flight mass spectrometry. Anal. Chem. 81, 6813–6822 (2009).
Stuart, T. & Satija, R. Integrative single-cell analysis. Nat. Rev. Genet. 20, 257–272 (2019).
Mereu, E. et al. Benchmarking single-cell RNA-sequencing protocols for cell atlas projects. Nat. Biotechnol. 38, 747–755 (2020).
Bassan, J. et al. TePhe, a tellurium-containing phenylalanine mimic, allows monitoring of protein synthesis in vivo with mass cytometry. Proc. Natl Acad. Sci. USA. 116, 8155–8160 (2019).
Rivello, F. et al. Single-cell intracellular epitope and transcript detection revealing signal transduction dynamics. Preprint at bioRxiv https://doi.org/10.1101/2020.12.02.408120 (2020).
Chung, H. et al. Simultaneous single cell measurements of intranuclear proteins and gene expression. Preprint at bioRxiv https://doi.org/10.1101/2021.01.18.427139 (2021).
Machado, L. et al. In situ fixation redefines quiescence and early activation of skeletal muscle stem cells. Cell Rep. 21, 1982–1993 (2017).
Simmons, A. J. et al. Cytometry-based single-cell analysis of intact epithelial signaling reveals MAPK activation divergent from TNF-α-induced apoptosis in vivo. Mol. Syst. Biol. 11, 835 (2015).
Macosko, E. Z. et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161, 1202–1214 (2015).
Klein, A. M. et al. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell 161, 1187–1201 (2015).
Angelo, M. et al. Multiplexed ion beam imaging of human breast tumors. Nat. Med. 20, 436–442 (2014).
Rodriques, S. G. et al. Slide-seq: a scalable technology for measuring genome-wide expression at high spatial resolution. Science 363, 1463–1467 (2019).
Vento-Tormo, R. et al. Single-cell reconstruction of the early maternal-fetal interface in humans. Nature 563, 347–353 (2018).
Efremova, M., Vento-Tormo, M., Teichmann, S. A. & Vento-Tormo, R. CellPhoneDB: inferring cell-cell communication from combined expression of multi-subunit ligand-receptor complexes. Nat. Protoc. 15, 1484–1506 (2020).
Browaeys, R., Saelens, W. & Saeys, Y. NicheNet: modeling intercellular communication by linking ligands to target genes. Nat. Methods 17, 159–162 (2020).
Jin, S. et al. Inference and analysis of cell-cell communication using CellChat. Nat. Commun. 12, 1088 (2021).
Miller, A. J. et al. Generation of lung organoids from human pluripotent stem cells in vitro. Nat. Protoc. 14, 518–540 (2019).
McCracken, K. W. et al. Modelling human development and disease in pluripotent stem-cell-derived gastric organoids. Nature 516, 400–404 (2014).
Boretto, M. et al. Development of organoids from mouse and human endometrium showing endometrial epithelium physiology and long-term expandability. Development 144, 1775–1786 (2017).
Driehuis, E., Kretzschmar, K. & Clevers, H. Establishment of patient-derived cancer organoids for drug-screening applications. Nat. Protoc. 15, 3380–3409 (2020).
Behbehani, G. K., Bendall, S. C., Clutter, M. R., Fantl, W. J. & Nolan, G. P. Single-cell mass cytometry adapted to measurements of the cell cycle. Cytom. A 81, 552–566 (2012).
Fienberg, H. G., Simonds, E. F., Fantl, W. J., Nolan, G. P. & Bodenmiller, B. A platinum-based covalent viability reagent for single-cell mass cytometry. Cytom. A 81, 467–475 (2012).
Bodenmiller, B. et al. Multiplexed mass cytometry profiling of cellular states perturbed by small-molecule regulators. Nat. Biotechnol. 30, 858–867 (2012).
Zunder, E. R. et al. Palladium-based mass tag cell barcoding with a doublet-filtering scheme and single-cell deconvolution algorithm. Nat. Protoc. 10, 316–333 (2015).
Willis, L. M. et al. Tellurium-based mass cytometry barcode for live and fixed cells. Cytom. A 93, 685–694 (2018).
McCarthy, R. L., Mak, D. H., Burks, J. K. & Barton, M. C. Rapid monoisotopic cisplatin based barcoding for multiplexed mass cytometry. Sci. Rep. 7, 3779 (2017).
Han, G., Spitzer, M. H., Bendall, S. C., Fantl, W. J. & Nolan, G. P. Metal-isotope-tagged monoclonal antibodies for high-dimensional mass cytometry. Nat. Protoc. 13, 2121–2148 (2018).
Finck, R. et al. Normalization of mass cytometry data with bead standards. Cytom. A 83, 483–494 (2013).
Cardoso, F., Qin, X. & Tape, C. J. TAPE-Lab/CyGNAL. https://zenodo.org/record/4849993 (2021).
McInnes, L. & Healy, J. UMAP: uniform manifold approximation and projection for dimension reduction. Preprint at https://arxiv.org/abs/1802.03426 (2018).
Levine, J. H. et al. Data-driven phenotypic dissection of AML reveals progenitor-like cells that correlate with prognosis. Cell 162, 184–197 (2015).
Orlova, D. Y. et al. Earth Mover’s Distance (EMD): a true metric for comparing biomarker expression levels in cell populations. PLoS One 11, e0151859 (2016).
Krishnaswamy, S. et al. Systems biology. Conditional density-based analysis of T cell signaling in single-cell data. Science 346, 1250689 (2014).
Dow, L. E. et al. Apc restoration promotes cellular differentiation and reestablishes crypt homeostasis in colorectal cancer. Cell 161, 1539–1552 (2015).
Gullaksen, S. E. et al. Titrating complex mass cytometry panels. Cytom. A 95, 792–796 (2019).
Takahashi, C. et al. Mass cytometry panel optimization through the designed distribution of signal interference. Cytom. A 91, 39–47 (2017).
Rein, I. D., Noto, H. O., Bostad, M., Huse, K. & Stokke, T. Cell cycle analysis and relevance for single-cell gating in mass cytometry. Cytom. A 97, 832–844 (2020).
Chevrier, S. et al. Compensation of signal spillover in suspension and imaging mass cytometry. Cell Syst. 6, 612–620.e5 (2018).
Trussart, M. et al. Removing unwanted variation with CytofRUV to integrate multiple CyTOF datasets. Elife 9, e59630 (2020).
van der Maaten, L. & Hinton, G. Visualizing data using t-SNE. J. Mach. Learn. Res. 9, 2579–2605 (2008).
Moon, K. R. et al. Visualizing structure and transitions in high-dimensional biological data. Nat. Biotechnol. 37, 1482–1492 (2019).
Acknowledgements
We are extremely grateful to L. Dow for sharing murine colonic organoids, M. Garnett and H. Francies for sharing PDOs, O. Ornatsky for providing 196cisplatin, S. Acton for providing murine tissue for fibroblast and macrophage isolation and A. Taylor and S. Guldin for OT-2 access and advice. We thank the UCL CI Flow-Core for CyTOF support. This work was supported by Cancer Research UK (C60693/A23783), the Cancer Research UK UCL Centre (C416/A25145), the Cancer Research UK City of London Centre (C7893/A26233), the UCLH Biomedical Research Centre (BRC422), The Royal Society (RSG\R1\180234) and The Rosetrees Trust (A1989).
Author information
Authors and Affiliations
Contributions
J.S. developed TOBis, designed rare earth metal-conjugated antibody panels and performed MC analysis. X.Q. designed and performed organoid and MC experiments, analyzed the data and wrote the manuscript. F.C.R. developed CyGNAL and wrote the manuscript. P.V. and M.R.Z. performed organoid and MC experiments. Y.J.B. and M.N. developed TeMal reagents. C.J.T. designed the study, analyzed the data and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
M.N. has pending intellectual property on the use of TeMal reagents for mass cytometry applications, which has been licensed to Fluidigm Corporation.
Additional information
Peer review information Nature Protocols thanks Kara L. Davis, Ozgun Gokce and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key reference using this protocol
Qin, X. et al. Nat. Methods 17, 335–342 (2020): https://doi.org/10.1038/s41592-020-0737-8
Supplementary information
Supplementary Information
Supplementary Figs. 1–6, Supplementary Tables 1 and 2 and Supplementary Method.
Supplementary Table 3
Debarcoding key for 35-plex TOBis
Supplementary Table 4
Debarcoding key for 126-plex TOBis
Rights and permissions
About this article
Cite this article
Sufi, J., Qin, X., Rodriguez, F.C. et al. Multiplexed single-cell analysis of organoid signaling networks. Nat Protoc 16, 4897–4918 (2021). https://doi.org/10.1038/s41596-021-00603-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-021-00603-4
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.