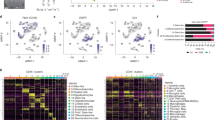
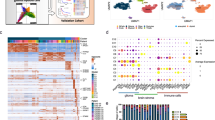
Abstract
Human tissue samples represent an invaluable source of information for the analysis of disease-specific cellular alterations and their variation between different pathologies. In cancer research, advancing a comprehensive understanding of the unique characteristics of individual tumor types and their microenvironment is of considerable importance for clinical translation. However, investigating human brain tumor tissue is challenging due to the often-limited availability of surgical specimens. Here we describe a multimodule integrated pipeline for the processing of freshly resected human brain tumor tissue and matched blood that enables analysis of the tumor microenvironment, with a particular focus on the tumor immune microenvironment (TIME). The protocol maximizes the information yield from limited tissue and includes both the preservation of bulk tissue, which can be performed within 1 h following surgical resection, as well as tissue dissociation for an in-depth characterization of individual TIME cell populations, which typically takes several hours depending on tissue quantity and further downstream processing. We also describe integrated modules for immunofluorescent staining of sectioned tissue, bulk tissue genomic analysis and fluorescence- or magnetic-activated cell sorting of digested tissue for subsequent culture or transcriptomic analysis by RNA sequencing. Applying this pipeline, we have previously described the overall TIME landscape across different human brain malignancies, and were able to delineate disease-specific alterations of tissue-resident versus recruited macrophage populations. This protocol will enable researchers to use this pipeline to address further research questions regarding the tumor microenvironment.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
All data generated or analyzed during this study are included in either this paper or our original research study5. Transcriptomic data generated using this pipeline are available at https://joycelab.shinyapps.io/braintime/. FCM data of the comparison of various brain tumor dissociation methods (included in Fig. 5a,b) have been deposited at the flow cytometry repository (http://flowrepository.org/): FR-FCM-Z3MF. Data not included in the aforementioned sources can be obtained from the corresponding author upon request due to patient privacy protection.
References
Achrol, A. S. et al. Brain metastases. Nat. Rev. Dis. Prim. 5, 5 (2019).
Stupp, R. et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 352, 987–996 (2005).
Quail, D. F. & Joyce, J. A. The microenvironmental landscape of brain tumors. Cancer Cell 31, 326–341 (2017).
Bejarano, L., Jordao, M. J. C. & Joyce, J. A. Therapeutic targeting of the tumor microenvironment. Cancer Discov. 11, 933–959 (2021).
Klemm, F. et al. Interrogation of the microenvironmental landscape in brain tumors reveals disease-specific alterations of immune cells. Cell 181, 1643–1660 e1617 (2020).
Friebel, E. et al. Single-cell mapping of human brain cancer reveals tumor-specific instruction of tissue-invading leukocytes. Cell 181, 1626–1642 e1620 (2020).
Long, G. V. et al. Combination nivolumab and ipilimumab or nivolumab alone in melanoma brain metastases: a multicentre randomised phase 2 study. Lancet Oncol. 19, 672–681 (2018).
Tawbi, H. A. et al. Combined nivolumab and ipilimumab in melanoma metastatic to the brain. N. Engl. J. Med. 379, 722–730 (2018).
Hendriks, L. E. L. et al. Outcome of patients with non-small cell lung cancer and brain metastases treated with checkpoint inhibitors. J. Thorac. Oncol. 14, 1244–1254 (2019).
Lim, M., Xia, Y., Bettegowda, C. & Weller, M. Current state of immunotherapy for glioblastoma. Nat. Rev. Clin. Oncol. 15, 422–442 (2018).
Schalper, K. A. et al. Neoadjuvant nivolumab modifies the tumor immune microenvironment in resectable glioblastoma. Nat. Med. 25, 470–476 (2019).
Bunevicius, A., Schregel, K., Sinkus, R., Golby, A. & Patz, S. REVIEW: MR elastography of brain tumors. Neuroimage Clin. 25, 102109 (2020).
Gritsenko, P. G. et al. p120-catenin-dependent collective brain infiltration by glioma cell networks. Nat. Cell Biol. 22, 97–107 (2020).
Leelatian, N. et al. Unsupervised machine learning reveals risk stratifying glioblastoma tumor cells. eLife https://doi.org/10.7554/eLife.56879 (2020).
Garofano, L. et al. Pathway-based classification of glioblastoma uncovers a mitochondrial subtype with therapeutic vulnerabilities. Nat. Cancer 2, 141–156 (2021).
Yuan, J. et al. Single-cell transcriptome analysis of lineage diversity in high-grade glioma. Genome Med. 10, 57 (2018).
Sankowski, R. et al. Mapping microglia states in the human brain through the integration of high-dimensional techniques. Nat. Neurosci. 22, 2098–2110 (2019).
Pombo Antunes, A. R. et al. Single-cell profiling of myeloid cells in glioblastoma across species and disease stage reveals macrophage competition and specialization. Nat. Neurosci. https://doi.org/10.1038/s41593-020-00789-y (2021).
Chen, A. X. et al. Single-cell characterization of macrophages in glioblastoma reveals MARCO as a mesenchymal pro-tumor marker. Genome Med. 13, 88 (2021).
Castellan, M. et al. Single-cell analyses reveal YAP/TAZ as regulators of stemness and cell plasticity in glioblastoma. Nat. Cancer 2, 174–188 (2021).
Richards, L. M. et al. Gradient of developmental and injury response transcriptional states defines functional vulnerabilities underpinning glioblastoma heterogeneity. Nat. Cancer 2, 157–173 (2021).
Kim, N. et al. Single-cell RNA sequencing demonstrates the molecular and cellular reprogramming of metastatic lung adenocarcinoma. Nat. Commun. 11, 2285 (2020).
Laughney, A. M. et al. Regenerative lineages and immune-mediated pruning in lung cancer metastasis. Nat. Med. 26, 259–269 (2020).
Rubio-Perez, C. et al. Immune cell profiling of the cerebrospinal fluid enables the characterization of the brain metastasis microenvironment. Nat. Commun. 12, 1503 (2021).
Berghoff, A. S. et al. Density of tumor-infiltrating lymphocytes correlates with extent of brain edema and overall survival time in patients with brain metastases. Oncoimmunology 5, e1057388 (2016).
Berghoff, A. S. et al. Correlation of immune phenotype with IDH mutation in diffuse glioma. Neuro Oncol. 19, 1460–1468 (2017).
Kohanbash, G. et al. Isocitrate dehydrogenase mutations suppress STAT1 and CD8+ T cell accumulation in gliomas. J. Clin. Invest. 127, 1425–1437 (2017).
Hodges, T. R. et al. Mutational burden, immune checkpoint expression, and mismatch repair in glioma: implications for immune checkpoint immunotherapy. Neuro Oncol. 19, 1047–1057 (2017).
Shih, D. J. H. et al. Genomic characterization of human brain metastases identifies drivers of metastatic lung adenocarcinoma. Nat. Genet. 52, 371–377 (2020).
Brastianos, P. K. et al. Genomic characterization of brain metastases reveals branched evolution and potential therapeutic targets. Cancer Discov. 5, 1164–1177 (2015).
Gromeier, M. et al. Very low mutation burden is a feature of inflamed recurrent glioblastomas responsive to cancer immunotherapy. Nat. Commun. 12, 352 (2021).
Berghoff, A. S. et al. Invasion patterns in brain metastases of solid cancers. Neuro Oncol. 15, 1664–1672 (2013).
O’Flanagan, C. H. et al. Dissociation of solid tumor tissues with cold active protease for single-cell RNA-seq minimizes conserved collagenase-associated stress responses. Genome Biol. 20, 210 (2019).
Hansen, D. M. et al. A holistic analysis of the intestinal stem cell niche network. Preprint at bioRxiv https://doi.org/10.1101/2019.12.12.871756 (2019).
Wenisch, C., Fladerer, P., Patruta, S., Krause, R. & Horl, W. Assessment of neutrophil function in patients with septic shock: comparison of methods. Clin. Diagn. Lab. Immunol. 8, 178–180 (2001).
Van Gassen, S., Gaudilliere, B., Angst, M. S., Saeys, Y. & Aghaeepour, N. CytoNorm: a normalization algorithm for cytometry data. Cytom. Part A 97, 268–278 (2020).
Plouffe, B. D., Murthy, S. K. & Lewis, L. H. Fundamentals and application of magnetic particles in cell isolation and enrichment: a review. Rep. Prog. Phys. 78, 016601 (2015).
Son, K. et al. Improved recovery of functionally active eosinophils and neutrophils using novel immunomagnetic technology. J. Immunol. Methods 449, 44–55 (2017).
Lozano-Ojalvo, D., et al. PBMC-derived T cells. in The Impact of Food Bioactives on Health: In Vitro and Ex Vivo Models (eds Verhoeckx, K. et al.) 169–180 (Springer, 2015).
Chometon, T. Q. et al. A protocol for rapid monocyte isolation and generation of singular human monocyte-derived dendritic cells. PLoS One 15, e0231132 (2020).
Ferrara, F. et al. Rapid purification of billions of circulating CD19+ B cells directly from leukophoresis samples. N. Biotechnol. 46, 14–21 (2018).
Swartzlander, D. B. et al. Concurrent cell type-specific isolation and profiling of mouse brains in inflammation and Alzheimer’s disease. JCI Insight https://doi.org/10.1172/jci.insight.121109 (2018).
National Society for Histotechnology (NSH). Guidelines for Hematoxylin & Eosin Staining. http://nsh.org/sites/default/files/Guidelines_For_Hematoxylin_and_Eosin_Staining.pdf (reviewed July 2001).
Picelli, S. et al. Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 9, 171–181 (2014).
Callahan, M. K., Williamson, P. & Schlegel, R. A. Surface expression of phosphatidylserine on macrophages is required for phagocytosis of apoptotic thymocytes. Cell Death Differ. 7, 645–653 (2000).
Macosko, E. Z. et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161, 1202–1214 (2015).
Acknowledgements
We express our gratitude to all patients who kindly agreed to donate tissue under protocol PB 2017-00240, F25/99. We thank the neurosurgery operating room staff and the technicians at the Pathology department of the Centre Hospitalier Universitaire Vaudois (CHUV) for their support in providing human patient samples; L. Bejarano Bosque, V. Wischnewski, A. Zomer and S. Watson in the Joyce lab for their technical assistance during sample processing; N. Piazzon for coordinating ATRX staining and IDH pyrosequencing performed at the Pathology department; the team of the UNIL Mouse Pathology Facility for cryosectioning of patient tissues; and K. Blackney and F. Sala de Oyanguren of the UNIL Flow Cytometry Facility for assistance with FAC sorting. Research in the Joyce lab is funded by the Carigest Foundation, ISREC Foundation, the Swiss Bridge Award, Breast Cancer Research Foundation, Cancer Research UK, Ludwig Institute for Cancer Research and the University of Lausanne. K.S. is supported in part by an Erwin-Schrödinger Fellowship from the Austrian Science Fund (FWF, J4343-B28). F.K. was supported in part by the German Research Foundation (DFG, KL2491/1-1) and Fondation Medic. A.A-P. is supported by an EMBO Long-term Postdoctoral Fellowship (EMBO ALTF 654-2019).
Author information
Authors and Affiliations
Contributions
F.K., R.L.B. and J.A.J. conceived the initial project; R.R.M., K.S., F.K., M.K. and R.L.B. designed and optimized pipeline modules; R.R.M., K.S., F.K., M.K., D.N.M. and A.A-P. performed experiments; R.R.M and K.S. analyzed data; R.B., D.L. and A.W. provided technical expertise for FCM experiments; R.B. and D.L. performed FAC sorting; J-P.B., R.D. and M.H. provided clinical material; J-P.B. performed histopathological review; R.R.M. and K.S. prepared the figures; R.R.M., K.S., F.K. and J.A.J. wrote the manuscript; J.A.J. supervised the project. All authors reviewed and edited the manuscript and approved the final draft.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Protocols thanks Burkhard Becher and the other, anonymous reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key reference using this protocol
Klemm, F. et al. Cell 181, 1643–1660.e17 (2020): https://doi.org/10.1016/j.cell.2020.05.007
Supplementary information
Supplementary Information
Supplementary Methods.
Rights and permissions
About this article
Cite this article
Maas, R.R., Soukup, K., Klemm, F. et al. An integrated pipeline for comprehensive analysis of immune cells in human brain tumor clinical samples. Nat Protoc 16, 4692–4721 (2021). https://doi.org/10.1038/s41596-021-00594-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-021-00594-2
This article is cited by
-
Phenotypic diversity of T cells in human primary and metastatic brain tumors revealed by multiomic interrogation
Nature Cancer (2023)
-
Clinical relevance of tumour-associated macrophages
Nature Reviews Clinical Oncology (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.