Abstract
As exemplified by the ongoing severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic, there is a strong demand for rapid high-throughput isolation pipelines to identify potent neutralizing antibodies for prevention and therapy of infectious diseases. However, despite substantial progress and extensive efforts, the identification and production of antigen-specific antibodies remains labor- and cost-intensive. We have advanced existing concepts to develop a highly efficient high-throughput protocol with proven application for the isolation of potent antigen-specific antibodies against human immunodeficiency virus 1, hepatitis C virus, human cytomegalovirus, Middle East respiratory syndrome coronavirus, SARS-CoV-2 and Ebola virus. It is based on computationally optimized multiplex primer sets (openPrimeR), which guarantee high coverage of even highly mutated immunoglobulin gene segments as well as on optimized antibody cloning and production strategies. Here, we provide the detailed protocol, which covers all critical steps from sample collection to antibody production within 12–14 d.
Similar content being viewed by others
Introduction
Monoclonal antibodies (mAbs) are essential molecules that are widely used for research, diagnostic and therapeutic applications. Over 90 mAbs have been approved by the US Food and Drug Administration, and mAbs have become one of the fastest-evolving markets in biotechnology1,2. Clinical applications of mAbs in infectious diseases are of special interest because of their (i) potential to target and neutralize infectious pathogens3,4,5,6, (ii) Fc-mediated effector functions7,8,9 and (iii) preferable pharmacokinetic profiles6,10,11,12. Moreover, by bridging innate and adaptive mechanisms, mAbs can act as immunomodulators that may stimulate the autologous immune response13,14,15. Substantial advancements in experimental techniques to identify and investigate antigen-specific B cells16,17,18,19 have paved the way to develop mAb-based prevention and treatment strategies20,21,22,23,24,25,26,27. In particular, the discovery of highly potent and broadly neutralizing mAbs demonstrates new opportunities for therapy and prophylaxis of infectious diseases28, as well as for structure-guided vaccination strategies29.
To date, several approaches have been described for the isolation of mAbs30. Notably, some of them allow the direct isolation of fully human antibodies from peripheral blood mononuclear cells (PBMCs) of infected or vaccinated individuals31,32,33,34,35,36,37,38, which circumvents the laborious and cost-intensive humanization of animal-derived mAbs39,40,41. In this protocol, we describe an optimized multiwell-format high-throughput antibody isolation pipeline that is highly reliable and effective, easy to use and covers the complete process from sample collection to antibody production.
Overview of the procedure
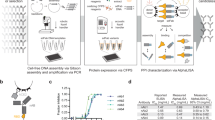
The described pipeline is built on single-cell antibody cloning workflows that were initially established by Küppers et al.16, Wardemann et al.17 and Tiller et al.42 and were further advanced over the past two decades19. The overall workflow comprises four basic stages: (I) B-cell isolation, (II) B-cell receptor (BCR) amplification and sequencing, (III) BCR cloning and (IV) recombinant antibody production (Fig. 1).
(I) Antigen-specific B-cell isolation
The first stage aims to isolate single B cells that specifically target the antigen of choice (Steps 1–30). To this end, single B cells are stained with fluorescently labeled antigens (e.g., proteins, virions or antigen-expressing cells) and sorted into multiwell plates (Fig. 1, stage I, Steps 18–30). We and others have successfully performed single B-cell sorting with numerous pathogen-derived surface proteins to identify pathogen-specific B cells34,36,37,38,43,44,45.
(II) BCR amplification
Next, a reverse transcription is performed, followed by amplification of naturally paired heavy and light chains in a nested or seminested multiprimer PCR (Fig. 1, stage II, Steps 31–69). Amplified heavy and light chains can then be analyzed by sequencing to identify clonal relationships and deduce repertoire characteristics from the isolated B cells (Steps 70–72). The choice of PCR primers is critical since they need to cover all different V gene segments, must not introduce mutations in the coding V regions and need to be compatible for multiplexing. To identify optimal multiplex primers for single BCR cloning, we recently developed openPrimeR, a program for designing and evaluating multiplex PCR primers for the amplification of highly diverse templates46,47. Using openPrimeR, we generated optimized primer sets for the amplification of all human IGH, IGK and IGL immunoglobulin V gene segments. These primer sets have been demonstrated to be preferable over previously published sets in terms of V gene coverage and the amplification of highly mutated antibody sequences36,47. In addition, we apply a single first-round multiplex PCR for the simultaneous preamplification of heavy- and light-chain sequences, which spares two complete PCR reactions in the process of antibody cloning37. Moreover, we scaled down all reactions of the nested PCR to half of the initial volumes, resulting in PCR cost reduction of 67% per cloned antibody37.
(III) BCR cloning
In the third stage, heavy- and light-chain PCR products are cloned into expression vectors (Fig. 1, stage III). To this end, a nested PCR is performed on the initial PCR products and cloning compatible overhangs are added (Steps 73–83). We modified our optimized multiplex primer sets to facilitate the convenient downstream assembly of individual variable regions into expression vectors by sequence- and ligation-independent cloning (SLIC)19 (Steps 95–117). Modified forward primers encode for the complete native leader sequence, whereas reverse primers were designed to target the conserved sequence motifs of the immunoglobulin constant region. This circumvents the design of individual V- and J-gene-specific cloning primers17,42. The modified SLIC primers have recently been used for the cloning of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) neutralizing antibodies, including one antibody that has entered clinical testing37.
(IV) Antibody production
Finally, antibodies are recombinantly produced for functional analysis, such as neutralizing activity (Fig. 1, stage IV). Recombinant antibody production has mainly been performed in large-volume suspension19,48 or large surface-adherent cell cultures18,42,49, which is space and cost consuming. We have therefore augmented the pipeline with a 48- or 96-well-format protocol for recombinant production of antibodies in adherent HEK293T cells (Step 135A(i–l)). Supernatants of transfected HEK293T cells typically yield microgram quantities of recombinant antibodies (as determined by human IgG capture ELISAs) and can be used without further purification for a number of downstream functional assays, such as antigen binding or virus neutralization. This allows for an efficient screening prior to large-scale production of a reduced number of selected candidate antibodies in suspension cell cultures, which yields milligram quantities for further characterization, such as cryogenic electron microscopy or in vivo studies34,36 (Step 135B(i–xxxv)).
Applications of the method
We have successfully used and validated the described approach for effective isolation of antigen-specific antibodies against human immunodeficiency virus 1 (HIV-1)36, human cytomegalovirus (HCMV)45, Ebola virus34 and SARS-CoV-237, as well as hepatitis C virus (HCV) and Middle East respiratory syndrome coronavirus (MERS-CoV, unpublished data). However, the pipeline is broadly applicable to a plethora of pathogens. Surface proteins of various viral pathogens have already been recombinantly expressed, e.g., for structural analyses50,51,52. Such surface proteins can be labeled with fluorophores and used for antigen-specific single B-cell sorting, as exemplified for respiratory syncytial virus, human papilloma virus, and Zika, Dengue and influenza virus43,44,53,54,55,56,57. Moreover, we also used this protocol to isolate Ebola-GP-specific B cells from immunized humans and HCMV gB-specific antibodies from humanized mice34,45.
Beside infectious diseases, the protocol may also be applicable to study the role of antigen-specific B cells and the respective BCR in a variety of different diseases. It may advance the understanding of autoantigen-specific B cells and their pathogenic potential in autoimmune disorders58,59,60 or help to investigate the role of B cells and the antibody repertoire in regulating antitumor immune responses61. Furthermore, the protocol is not limited to studies of antigen-experienced memory B cells but can be exploited to study the BCR of other B-cell subsets such as transitional or mature-naive B cells. Taken together, this modified pipeline is of great value to study B-cell and antibody responses against a wide range of antigenic structures in a short amount of time.
Comparison with other methods
In the past, several approaches and combinations of different protocols have been developed to analyze B-cell repertoires and generate mAbs. These include bait protein-based cell sorting and antibody cloning techniques that were both established and advanced in the Nussenzweig Laboratory18,19,36,37,38,42,43,55,62, as well as panning of combinatorial display libraries63,64, and supernatant screenings of single B-cell cultures32 or immortalized B cells65. These approaches have been successfully applied to identify numerous antibody candidates for research, diagnostics and therapy16,17,36,37,54,66,67.
Combinatorial display libraries enable the rapid generation of large amounts of different antibodies. However, they also have critical drawbacks. First, heavy and light chains are randomly paired and expressed in combinations that might not naturally occur, whereas some cognate pairings might not be generated63,64,68. Second, multiple rounds of selection can lead to enrichment of clones that do not necessarily exhibit high binding but are rather expressed and displayed more efficiently. As a consequence, rare but desirable clones might get lost and isolated antibodies might not represent the natural clonal distribution. In contrast, isolation and analysis of individual B cells, as included in the described pipeline, preserve the natural pairing of heavy and light chains and, thus, natural clonality.
Single-cell isolation steps are also performed in approaches that rely on the prescreening of supernatants, e.g., after immortalization33,69 or single-cell culture35,70. However, supernatant prescreening requires the cost- and time-consuming cultivation of thousands of B cells71, as exemplified by the pan-influenza A neutralizing antibody F16, for which over 100,000 B cells were cultivated and screened72. This method is therefore less appropriate for the high-throughput isolation of rare antibodies, such as those derived from antigen-specific memory B cells, which typically comprise less than 0.5% of the memory IgG compartment34,36,37,73. Therefore antigen-specific single B-cell sorting is often advantageous in successfully isolating novel mAbs36,37,38,43,44,56,66,74,75.
PCR coverage and primer formulations can represent a bottleneck in current PCR-based antibody cloning protocols, as they critically affect amplification efficiency of the variable regions of the immunoglobulin loci, as well as costs, scalability and simplicity of the workflow. Most B-cell isolation pipelines report mean PCR amplification efficiencies of 50–74% for heavy- and 40–65% for light-chain sequences, respectively18,19,38,42. On average, 30–65% of paired heavy- and light-chain sequences are recovered from single-cell PCRs38,42. By contrast, the protocol proposed here yields recoveries of >89% for paired heavy and light chains. This high recovery is the prerequisite for a high throughput capacity. It allows a direct propagation of the initial sorting layout throughout the whole process, sparing time-consuming compilations of positive PCR hits into new 96-well plates. In addition, it guarantees an efficient consumption of reagents and facilitates the cost-effective use of 96-well-format PCR purification kits for cloning PCRs. Of note, our optimized primers are particularly suitable for amplifying highly mutated antibody sequences47. Importantly, by adapting all three PCR steps in the pipeline to the stringent use of the optimized multiplex primer mixes, we also simplified the generation of SLIC-compatible PCR products, allowing a fast and reliable cloning of mAbs in a 96-well format.
Current protocols are often limited in sample throughput by subsequent antibody expression in high volume suspension or large tissue cultures18,19,42. We therefore adapted the pipeline to a 48- or 96-well-plate format for antibody expression. Despite substantially increased sample throughput, the described protocol still provides comparable transfection efficiencies and antibody concentrations to existing workflows18,19,38,42. Approximately 85–90% of transfections produce recombinant mAbs with concentrations ≥1 µg/ml (mean 10 µg/ml, median 6.4 µg/ml) and yields >0.4 µg (mean 4.2 µg, median 2.6 µg) in a 48-well format (mean/median concentration of 32 µg/ml or 31 µg/ml and mean/median yield of 6.3 µg and 6.2 µg in a 96-well format). In addition, with as little as 12–14 d for its completion, the described pipeline ranks among the fastest currently published pipelines for the isolation of antigen-specific antibodies19,38.
Experimental design
To simplify the parallel processing of many cDNA plates for high-throughput production, we adapted all protocols of the pipeline to a stringent use of multiwell plates, including the incorporation of 96-well plate format purification steps for cloning PCR products, mini-scale DNA preparations and glycerol stock storage. The detailed experimental workflow is depicted in Fig. 2.
For antigen-specific B-cell isolation (I, days 1 and 2), PBMCs are isolated by density centrifugation from peripheral blood and CD19+ B cells are magnetically enriched before single-cell sorting with a fluorescently labeled bait protein into 96-well plates. For highly efficient BCR amplification and cloning (II and III, days 3–7), mRNAs of individual B cells are reversed transcribed and variable regions are preamplified with computationally optimized primer mixes in a multiplex PCR (first). This first PCR product serves as a template for seminested chain-specific enrichment PCRs (second), which are analyzed by Sanger sequencing. For cloning (stage III), first PCR products are amplified with computationally optimized primer mixes in a cloning PCR (third) to add overhangs for subsequent SLIC into mammalian expression vectors and transformation into E. coli. For each chain, individual clones are picked and used to inoculate both a 96-well plate liquid culture and a colony PCR reaction mix. The colony PCR is analyzed by sequencing to identify correctly inserted variable regions and the liquid culture can be supplemented with glycerol for long-term storage. For high-throughput and/or large-scale antibody production (IV, days 8–12/14), either the liquid cultures or glycerol stocks of correct clones are used for inoculation of 96-well mini-scale (high-throughput) or shaking flask midi-scale (large-scale, low-throughput) bacterial cultures. For the high-throughput protocol, DNA is prepared with 96-well-format plasmid-DNA isolation columns, and HEK293T cells are transfected in a 48- or 96-well format. Supernatants can be directly used without further purification for downstream assays such as ELISA or microneutralization assays. For large-scale production, HEK293-6E cells are transfected with midi-scale DNA preparations of heavy and light chains and mAbs are purified by protein G affinity purification for downstream assays.
(I) Antigen-specific B-cell isolation
Any human sample collection must (i) be performed in accordance with a study protocol, (ii) be approved by the local institutional review board (IRB) and (iii) have signed informed consent from all study participants. As an alternative to a blood draw, large numbers of leukocytes can be collected by leukapheresis if comprehensive and systemic investigation of B cells is limited by cell numbers and the volume of blood that can be drawn safely from study participants. Leukapheresis is a well-tolerated and safe procedure that is routinely performed as an outpatient procedure in 2–4 h and typically yields 4–12 × 109 PBMCs with only minimal loss of erythrocytes. Subsequent to sample collection, PBMCs are isolated by standard density gradient centrifugation using Histopaque separation medium and Leucosep cell tubes. Cell count as well as viability are determined, and PBMCs are subjected either to cryopreservation or to B-cell isolation. From the purification of 1 ml of whole blood, isolation of ~1 × 106 PBMCs with a cell viability ≥90% can be expected. CD19+ B cells are positively selected from fresh or frozen PBMCs by magnetic-activated cell sorting (MACS), and cell count and viability are determined. Among PBMCs, we expect to select 5–10% B cells with a viability of ≥80%. For single-cell sorting, CD19+ B cells are stained with anti-CD20, anti-IgG (class-switched memory B cells), 4′,6-diamidin-2-phenylindol (DAPI, viability marker) and the respective bait protein for the selection of antigen-specific B cells. Importantly, bait proteins should be tested for unspecific staining using appropriate negative controls. The gating strategy includes the identification of lymphocytes, exclusion of cell doublets and dead cells, and the selection of IgG+ B cells. The final gate for sorting is set on IgG+ B cells binding to the pathogen-specific bait protein. To exclude unspecific background staining and to improve the specificity of the sorting gate, IgG+ B cells can also be double-stained with two bait proteins labeled with different fluorochromes. Single antigen-specific B cells are sorted into separate wells of a 96-well plate containing sorting buffer. For negative controls of subsequent RT-PCR reactions, we recommend sparing 5–10% of wells per 96-well plate.
(II) High-efficiency BCR amplification
cDNA is generated from single B-cell RNA by reverse transcription with random hexamer primers. Heavy- and light-chain variable (V) regions are then amplified by seminested multiplex PCR. To this end, a single first-round multiplex PCR is performed with optimized primer sets that target the 5′ leader region and the immunoglobulin constant region, followed by three separate second-round PCRs for the targeted amplification of heavy and light chains with nested reverse primers. Aliquots of the second PCR products are analyzed by agarose gel electrophoresis for the presence of the correct product at ~500 bp for heavy- and ~450 bp for light-chain sequences, respectively (coverages should exceed 90% for the amplification of IgG heavy and 85% for kappa and lambda light chains). Positive hits are analyzed by Sanger sequencing and annotated with IgBlast76 (according to the IMGT system77) to investigate repertoire characteristics and to define clonal relations of antibody sequences. Methods and criteria for clonal assignment typically vary between different laboratories and publications. For example, IgBLAST defines clonotypes as sequences with the same V, D and J gene as well as identical CDR3 nucleotide sequences76. We apply less stringent criteria to account for somatic hypermutation as well as sequencing or PCR errors within the CDR3. To this end, we usually group sequences by identical V and J genes with a maximum of one amino acid difference in CDRH3 length and a total CDRH3 difference of ≤25% on the amino acid level. Depending on time and resources, cloning can be continued with the first PCR products from all sequenced antibodies or from preselected antibodies on a compilation plate.
(III) High-efficiency BCR cloning
Selected heavy- and light-chain variable regions are cloned into mammalian antibody expression vectors encoding the human IgG1 heavy-, lambda- or kappa light-chain constant region42 by SLIC19. To this end, expression vectors are linearized and high-fidelity PCRs on the first PCR products are performed with optimized SLIC primers that add a complete endogenous secretion signal for expression as well as SLIC-compatible overhangs. Negative controls should be included in the cloning PCR to exclude contaminations. PCR products are analyzed by agarose gel electrophoresis for product lengths of ~600 bp for heavy and ~550 bp for light chains. Efficiencies of up to 90% for the amplification of heavy and kappa or up to 85% for lambda light chains can be expected. For a higher recovery of lambda light chains, they can be amplified in a separate first single-cell PCR prior to cloning PCR (described in Troubleshooting). PCR products are then purified with 96-well-format silica membranes, cloned into linearized expression plasmids with T4 DNA polymerase (i.e., by SLIC) and transformed into Escherichia coli. Individual clones are picked to inoculate liquid cultures, and the correct insertion of the heavy- or light-chain V region sequence into the expression vector is examined by low-fidelity (Taq-based) colony PCR. To identify correct clones for antibody expression, sequences of the colony PCR products are aligned to the corresponding second PCR sequences. Besides sequence homology, (i) the presence of a start codon and a complete leader sequence, (ii) the correct reading frame at the junction between FWR4 and the constant region and (iii) the absence of stop codons should be checked. This process can be performed manually with commercial alignment software or automatically using scripting languages such as Python. Finally, mini- or midi-scale DNA preparations are inoculated from the liquid cultures for each correct pair of heavy- and light-chain sequences.
(IV) High-throughput and/or large-scale antibody production
After insertion of variable region sequences into expression vectors, the subsequent experimental procedures can be conducted either as a high- or low-throughput approach. Low- and high-throughput approaches differ in their primary objectives and downstream applications. The high-throughput approach allows for the rapid microgram-scale isolation of hundreds of different antibodies in a 48- or 96-well format. It relies on mini-scale DNA preparations and chemical transfection of adherent HEK293T cells. Supernatants of transfected cells can directly be used for characterization in microneutralization assays or binding ELISAs. Alternatively, antibodies can be produced in milligram-scales, following a low-throughput approach, which relies on midi-scale DNA preparations, cationic polymer transfection of 50 ml suspension cell cultures and protein G affinity purification. This approach yields enough purified recombinant antibodies for more-consuming downstream experiments such as half-life determination in small animal models, autoreactivity assays or structural analyses.
-
(A)
High-throughput antibody production. For 48-well transfection, 6 × 104 HEK293T cells/well are seeded in starving medium 18–24 h prior to transfection (4 × 104 cells/well for 96-well format). Higher cell counts or overgrowth of cell cultures might substantially reduce transfection efficiencies. HEK293T cells of each well are cotransfected with the heavy- and corresponding light-chain expression vectors and incubated for 96 h. Transfection efficiency is controlled by transfection of a GFP-encoding plasmid in a control well, which can be detected by fluorescence microscopy 24 h after transfection. Supernatants of HEK293T cells are harvested 96 h after transfection and concentrations of produced antibodies are measured by human IgG capture sandwich ELISA. Supernatants of HEK293T cells can directly be used for downstream characterization assays without further purification.
-
(B)
Large-scale antibody production. Depending on the primary experimental objective and required yield of purified antibodies, 8 × 105 to 1.2 × 106 HEK293-6E cells per ml are transfected in a culture volume of at least 50 ml. HEK293-6E cells are cotransfected with heavy- and light-chain expression plasmids and supernatants are harvested 7 d after incubation under constant shaking in a cell incubator. Recombinant antibodies can be used for downstream assays after protein G–based affinity purification.
Expertise and specialized equipment needed to implement the protocol
Informed consent from human blood donors must be obtained and peripheral blood draws must be performed by trained healthcare personnel. Isolation of single antigen-specific B cells requires a FACS core facility equipped with a suitable cell sorter and trained laboratory staff who are experienced in operating and setting up the FACS device. To conduct the complete workflow within the stated timeframe, Sanger sequencing of crude PCR products needs to be performed overnight. We have outsourced this step to a globally acting sequencing company that provides DNA cleanup and sequencing of 96-well plates within one working day. The rest of the pipeline requires standard molecular biology and cell-culture laboratories but no further specialized equipment. However, since single-cell RT-PCRs are highly susceptible to nucleic acid contaminations, it is strongly recommended perform that single-cell RT-PCRs be performed in a designated and separated single-cell PCR room or working area, where experiments with other DNA templates, such as antibody-encoding plasmids, are strictly prohibited. Master mixes for single-cell RT-PCRs should exclusively be prepared inside a designated PCR cabinet, which should be regularly decontaminated by UV irradiation and DNA decontaminating reagents. For maximum throughput of the wet-lab protocol, we advise having access to at least three (optimally six or more) 96-well thermocyclers and including one or two experienced PhD students or postdocs. We highly recommend automating computational analyses with scripting language, which requires advanced bioinformatics expertise. However, all analyses can be performed on standard desktop computers with minimal software requirements (including Microsoft Excel or comparable spreadsheet software, as well as an internet connection to access annotation online tools) and require only a basic level of bioinformatics expertise.
Limitations
The described experimental pipeline relies on a preselection of single antigen-specific B cells by fluorescently labeled antigens in the form of recombinant proteins, virions or whole cells. For adapting this protocol to a specific scientific question, the choice of the bait protein should be carefully considered, as it will critically determine the selection of antibodies. For example, isolation efficiencies for broadly neutralizing antibodies targeting certain structure-dependent epitopes of the HIV-1 envelope strongly differ between BG505 SOSIP.664 gp140-trimers versus monomeric YU2 gp120 as bait proteins50. Consequently, B cells that are reactive to immunogenic structures not present on the chosen bait (e.g., certain conformational epitopes or protein subunits) will be excluded from the isolation. Furthermore, generation of fluorescently labeled antigens in their native configuration is often complicated by substantial challenges in isolation, expression or stabilization of soluble bait proteins50,78.
In most instances, the described pipeline is limited by the available equipment (i.e., mainly the number of thermocyclers, centrifuges, gel-electrophoresis systems and incubators) as well as human resources. The first rate-limiting step in the pipeline is the second PCR (Steps 57–69), where three individual reactions (i.e., heavy-, kappa- and lambda-chain amplification) have to be performed for each initial B cell. Given access to six 96-well thermocyclers and one large electrophoresis gel system, 12 second PCR runs and analyses can be performed in 1 d, if properly scheduled. The second rate-limiting step is the SLIC reaction, where each transformation (i.e., one heavy and one light per antibody) needs to be streaked out on a Petri dish (Steps 95–117). The third and last rate-limiting step is the colony PCR (Steps 118–134), where three clones per transformation are analyzed (i.e., six PCR reactions per antibody, three for each chain).
Taken together, a single experimenter can process at least two sorted 96-well plates (i.e., 186 antibodies, if three wells are left empty as negative controls) in a single run. However, there will be many steps where more samples can be handled in parallel. For example, a single experimenter can process 372 B cells (i.e., four sorted 96 plates, with three empty negative controls per plate) during reverse transcription (Steps 31–47) and first PCR (Steps 48–56) in parallel. For implementation it is recommended that the whole pipeline be run with a single 96-well plate of sorted cells and then start to scale up. With the modular structure of the individual stages and careful scheduling, several researchers can work in parallel on different steps, an approach that enables sequencing the BCR of over 4,000 B cells within less than 2 months37. The throughput can be substantially increased when transferring the protocol to a liquid handling robot and switching to 384-well plates.
Materials
Reagents
-
0.5 M EDTA (Thermo Fisher Scientific, cat. no. AM9260G)
-
6× DNA loading dye (Thermo Fisher Scientific, cat. no. R0611)
-
ABTS substrate solution (Thermo Fisher Scientific, cat. no. 002024)
-
AbVec1.1-IGKC (IgK) expression vector (European Nucleotide Archive, accession no. LT615369)
-
AbVec1.1-IGLC2-XhoI (IgL) expression vector (Addgene, accession no. 99575)
-
AbVec2.0-IGHG1 (IgG1) expression vector (European Nucleotide Archive, accession no. LT615368)
-
Acetic acid 100% (Carl Roth, cat. no. 6755.2)
Caution
Acetic acid is highly corrosive to eyes, mucous membranes and skin. Always handle acetic acid with gloves and splash goggles.
-
Agar, bacteriology grade (PanReac AppliChem, cat. no. A0949)
-
Agarose Basic (PanReac AppliChem, cat. no. A8963)
-
Alexa Fluor 700 mouse anti-human CD20 (BD biosciences, cat. no. 560631)
-
Alkaline phosphatase, calf intestinal phosphatase (CIP; New England BioLabs, cat. no. M0290S)
-
Ampicillin sodium salt (Sigma Aldrich, cat. no. A0166)
-
Antibiotic–antimykotic (100×) (Thermo Fisher Scientific, cat. no. 15240062)
-
APC mouse anti-human IgG (BD Biosciences, cat. no. 550931)
-
BG505 SOSIP.664 gp140 (ref. 50)
-
BirA500: BirA biotin-protein ligase standard reaction kit (Avidity, cat. no. BirA500)
-
Bovine serum albumin (BSA; Carl Roth, cat. no. 8076.3)
-
BsiWI-HF restriction enzyme (New England BioLabs, cat. no. R3553L)
-
CD19 microbeads, human (Miltenyi Biotec, cat. no. 130-050-301)
-
Chemically competent E. coli, Library Efficiency DH5⍺ Competent Cells (Thermo Fisher Scientific, cat. no. 18263012)
-
CutSmart Buffer (New England BioLabs, cat. no B7204S)
-
4′,6-Diamidino-2-phenylindole, dihydrochloride (DAPI) (Thermo Fisher Scientific, cat. no. D1306)
-
Dipotassiumhydrogenphosphate (K2HPO4; Carl Roth, cat. no 6875.1)
-
Disodiumhydrogenphosphate (Na2HPO4; Carl Roth, cat. no. 7558-79-4)
-
Dimethyl sulfoxide, Hybrid-Max (Sigma Aldrich, cat. no. D2650-100ml)
-
DL-dithiothreitol (DTT) (Promega, cat. no. P1171)
-
DNA AWAY (Carl Roth, cat. no. X996.1)
-
DyLight 488 NHS Ester (Thermo Fisher Scientific, cat. no. 46403)
-
DyLight 650 NHS Ester (Thermo Fisher Scientific, cat. no. 62266)
-
dNTP mix, 25 mM (Thermo Fisher Scientific, cat. no. R1122)
-
Dulbecco’s Modified Eagle Medium (DMEM; Gibco, cat. no. 11960-044)
-
Dulbecco’s phosphate-buffered saline (DPBS) (Thermo Fisher Scientific, cat. no. 14190-169)
-
EBOV GPΔTM34
-
EcoRI-HF restriction enzyme (New England BioLabs, cat. no. R3101S)
-
EDTA disodium salt, dihydrate (Carl Roth, cat. no. 8043.2)
-
Ethanol 99.8% Rotipuran (Carl Roth, cat. no. 9065.5)
-
Fetal bovine serum (FBS; Sigma Aldrich, cat. no. F9665)
-
FITC mouse anti-human IgM (BD Biosciences, cat. no. 555782)
-
FreeStyle293 expression medium (Thermo Fisher Scientific, cat. no. 12338026)
-
GeneRuler 1 kb Plus DNA Ladder (Thermo Fisher Scientific, cat. no. SM1331)
-
Glycerol (Carl Roth, cat. no. 7530.1)
-
Glycine (Carl Roth, cat. no. 3187.3)
-
Hank’s balanced salt solution (Sigma Aldrich, cat. no. H6648)
-
HEK293-6E suspension cells (National Research Council of Canada, NRC file 11565)
Caution
Check cell lines regularly for Mycoplasma infection.
-
HEK293T adherent cells (ATCC, cat. no. CRL-11268)
-
Heparin sodium 25,000 IE/5 ml (B. Braun, cat. no. 2047217)
-
Histopaque-1077 Hybri-Max 500 ml (Sigma Aldrich, cat. no. H8889-500ml)
-
HRP-conjugated polyclonal goat anti-human IgG, Fcγ Fragment Specific (Jackson ImmunoResearch, cat. no. 109-035-098)
-
Hydrochloric acid (Carl Roth, cat. no. T134.1)
-
IgG1 kappa from human myeloma plasma (Sigma Aldrich, cat. no. I5154)
-
l-Glutamine, 200 mM (Gibco, cat. no. 25030081)
-
NP-40 Surfact-Amps Detergent Solution (Thermo Fisher Scientific, cat. no. 85124)
-
Nuclease-free water, not DEPC-treated (Thermo Fisher Scientific, cat. no. AM9937)
-
PE streptavidin (BD Biosciences, cat. no. 349023)
-
Phosphate-buffered saline (PBS) pH 7.4, 1× (Gibco, cat. no. 10010056)
-
PBS pH 7.4, 10×, RNase-free (Thermo Fisher Scientific, cat. no. AM9624
-
Platinum Taq DNA Polymerase (Thermo Fisher Scientific, cat. no. 10966026)
-
Platinum Taq Green Hot Start DNA Polymerase (Thermo Fisher Scientific, cat. no. 11966034)
-
Polyclonal goat anti-human IgG, Fcγ Fragment Specific (Jackson ImmunoResearch, cat. no. 109-005-098)
-
Polyethylenimine (PEI), linear (Polyscience, cat. no. 23966-2)
Caution
PEI is a skin and eye irritant. Handle PEI with gloves and inside a hood with laminar flow.
-
Potassium chloride (Thermo Fisher Scientific, cat. no. AM9640G)
-
Potassiumdihydrogenphosphate (KH2PO4; Carl Roth, cat. no. 3904.1)
-
Protein G Sepharose 4 Fast Flow (Sigma Aldrich, cat. no. GE17-0618-05)
-
pWPXL EGFP expressing vector (Addgene, cat. no. 12257)
-
Q5 Hot Start High-Fidelity DNA Polymerase (New England BioLabs, cat. no. M0493L)
-
Random hexamer primers 200 ng/µl (Thermo Fisher Scientific cat. no. S0142)
-
Recombinant RNasin ribonuclease inhibitor (Promega, cat. no. N2515)
-
RNase AWAY (Carl Roth, cat. no. A998.3)
-
RNaseOUT recombinant ribonuclease inhibitor (Invitrogen, cat. no. 10777019)
-
RPMI 1640 medium (Thermo Fisher Scientific, cat. no. 61870044)
-
SalI-HF restriction enzyme (New England BioLabs, cat. no. R3138S)
-
SOC Medium (Thermo Fisher Scientific, cat. no. 15544034)
-
Sodium chloride (salt; Carl Roth, cat. no. 3957.2)
-
Sodium pyruvate, 100 mM (Gibco, cat. no. 11360070)
-
SuperScript IV Reverse Transcriptase (Thermo Fisher Scientific, cat. no. 18090200)
-
SYBR Safe DNA Gel Stain (Thermo Fisher Scientific, cat. no. S33102)
-
T4 DNA Polymerase (New England BioLabs, cat. no. M0203S)
-
Tris-(hydroxymethyl)-aminomethane (Carl Roth, cat. no. 4855.3)
-
Trypan blue solution (Sigma Aldrich, cat. no. T8154)
Caution
Trypan blue might develop turbidity or sediment during storage.
-
Trypsin–EDTA, 0.05% (Thermo Fisher Scientific, cat. no. 25300054)
-
Tryptone (Carl Roth, cat. no. 8952.2)
-
Turbofect transfection reagent (Thermo Fisher Scientific, cat. no. R0531)
-
Tween-20 (Carl Roth, cat. no. 9127.2)
-
XhoI restriction enzyme (New England BioLabs, cat. no. R0146L)
-
Yeast extract (Carl Roth, cat. no. 2363.2)
-
YU2gp140 (ref. 62)
Equipment
-
10 ml serological pipette (Sarstedt, cat. no. 861254001)
-
10 µl filter tips (Sarstedt, cat. no. 70.1130.210)
-
1,000 µl filter tips (Sarstedt, cat. no. 70.762.211)
-
15 ml plastic tube (Sarstedt, cat. no. 62.554.502)
-
200 µl filter tips (Sarstedt, cat. no. 70.760.213)
-
25 ml serological pipette (Sarstedt, cat. no. 86.1685.00)
-
5 ml round-bottom polystyrene test tube with cell strainer snap cap (Corning, cat. no. 734-0001)
-
5 ml serological pipette (Sarstedt, cat. no. 86.1253.00)
-
5 ml tube with snap cap (VWR cat. no. 525-0796)
-
50 ml plastic tube (Sarstedt, cat. no. 62.547.254)
-
50 ml serological pipette (Sarstedt, cat. no. 86.1256.001)
-
50 ml syringe (B. Braun, cat. no. 4617509F)
-
Adhesive PCR plate foil (Thermo Fisher Scientific, cat. no. AB0626)
-
Amicon Ultra-4, 30 kDa tube (Merck Millipore, cat. no. UFC803096)
-
BD FACS Aria Fusion (BD biosciences)
-
Bench top centrifuge 5424 R (Eppendorf, cat. no. 5404000010)
-
Blue light transilluminator, ECX-F20.SkyLight (VWR, cat. no. 732-1359DE)
-
C-chip Neubauer improved (Carl Roth, cat. no. T729.1)
-
Cell culture flasks, 250 ml (Corning, cat. no. CLS431407)
-
Cell culture plate, 96-well, surface: standard, flat base (Sarstedt, cat. no. 83.3924.500)
-
Chromatography columns (BioRad, cat. no. 7311550)
-
CO2 cell incubator (Panasonic, cat. no. MCO-230AIC)
-
CO2 cell incubator with shaker New Brunswick S41i (Eppendorf, cat. no. S41l230011)
-
Costar 96-well ELISA plates EasyWash, HighBinding (Corning, cat. no. 3369)
-
CryoPure vial, 1.8 ml (Sarstedt, cat. no. 72.379.007)
-
Dermagrip nitril, latex-free glove (Remesco, cat. no. D1502-17)
-
Dispenser tip, 25 ml (VWR, cat. no. 613-2065)
-
Electrophoresis power supply (Life Technologies, cat. no. PS0091)
-
Electrophoresis system owl A3-1 (Thermo Fisher Scientific, cat. no. A3-1)
-
Eppendorf 5810R centrifuge, rotor S-4-104) (Thermo Fisher Scientific, cat. no. 12836213)
-
Feather, scalpel no. 21 (Pfm medical, cat. no. 200210021)
-
Flap cap strips, colorless (Peqlab, cat. no. 82-0784-A)
-
Gel Doc XR+ Gel documentation system (Bio-Rad, cat. no. 5838)
-
Heavy duty labels (Avery Zweckform, cat. no. L6008-20)
-
Hydrospeed ELISA plate washer (Tecan, cat. no n/a)
-
Incubation shaker (Infors HT, cat. no. n/a)
-
Incubator (Binder, cat. no. 9010-0081)
-
Inoculation loop (Carl Roth, cat. no. EA90.1)
-
Leucosep centrifuge tube (Greiner Bio-One, cat. no. 227290P)
-
MACS LS columns (Miltenyi Biotec, cat. no. 130-042-401)
-
Microtest Plate 96-well (Sarstedt, cat. no. 82.1583)
-
Mini centrifuge (neoLab, cat. no. D-8850)
-
Multichannel pipette 0.5–10 µl (VWR, cat. no. 613-0884)
-
Multichannel pipette 10–100 µl (VWR, cat. no. 613-5904)
-
Multidispenser pipette (Eppendorf, cat. no. 4982000012)
-
NanoDrop One spectral photometer (Thermo Fisher Scientific, cat. no. ND-ONE-W)
-
NucleoBond Xtra Midi (Macherey Nagel, cat. no. 740410.100)
-
NucleoSpin 96 PCR Clean-up Kit (Macherey Nagel, cat. no. 740658.4)
-
NucleoSpin 96 Plasmid (Macherey Nagel, cat. no. 740625.4)
-
NucleoSpin Gel and PCR Clean-up (Macherey Nagel, cat. no. 740609.250)
-
Nunc 96 DeepWell plate, nontreated (Sigma Aldrich, cat. no. Z688738-32EA)
-
Parafilm M (Carl Roth, cat. no. H666.1)
-
PCR plate half skirt, 96-well (Sarstedt, cat. no. 72.1979.102)
-
Petridish 60 × 15 mm (Sarstedt, cat. no. 82.1194.500)
-
Plate reader sunrise (Tecan, cat. no. n/a)
-
Preseparation filter (70 µm) (Miltenyi BioTec, cat. no. 130-095-823)
-
Quadro MACS Separator (Miltenyi BioTec, cat. no. 130-090-976)
-
Safe-Lock Tube, 1.5 ml (Sarstedt, cat. no. 0030120.086)
-
Single-use reagent pipetting reservoir, 100 ml (Carl Roth, cat. no. EKX3.1)
-
Single-use reagent pipetting reservoir, 25 ml (Carl Roth, cat. no EKT7.1)
-
Strata cooler cryopreservation module (Aligent Technologies, cat. no. 50-125-551)
-
Thermomixer (Eppendorf, cat. no. 5384000012)
-
Cell culture plate, 48-well, surface: standard, flat base (Sarstedt, cat. no. 83.3923.005)
-
Transferpette 20–200 µl (Brand, cat. no. 705930)
-
Transferpette 30–300 µl (Brand, cat. no. 705932)
-
Tube 50 ml, 114 × 28 mm, PP (Sarstedt, cat. no. 62.547.254)
-
Tube rotator (Stuart, cat. no. SB3)
-
High-speed centrifuge (Beckmann Coulter, cat. no. B22987)
-
Ultrafree-CL filter columns, 0.22 µm (Merck Millipore, cat. no. UFC40GV0S)
-
Ultra-low-temperature freezer (−150 °C) (Panasonic, cat. no. MDF-C2156VAN)
-
Ultra-low-temperature freezer (−80 °C) (Panasonic, cat. no. MDF-DU700VH)
-
Veriti 96-well thermal cycler (Thermo Fisher Scientific, cat. no. 4375786)
-
Vortex mixer (neoLab, cat. no. D-8900)
Reagent setup
Tris-acetate-EDTA buffer (TAE buffer) (50×)
Dissolve 242 g of Tris-(hydroxymethyl)-aminomethane in 600 ml of distilled H2O. Add 200 ml of 0.5 M EDTA (pH 8.0) and 57.1 ml of acetic acid. Fill up to a final volume of 1 L with distilled H2O. Autoclave and store the buffer at room temperature (20–24 °C) for up to 5 years.
FACS buffer
Add 10 ml of FBS and 2 ml of 0.5 M EDTA to 488 ml of 1× PBS. Store FACS buffer at 4–8 °C for up to 3 months.
Freezing medium (vol/vol)
Medium consists of 90% FBS + 10% DMSO and can be stored for 1 d at 4 °C. Cool freezing medium to 4 °C prior to usage.
MACS buffer
Add 200 µl of 0.5 M EDTA and 5 ml of 5% (wt/vol) BSA to 45 ml of 1× PBS. MACS buffer can be stored at 4–8 °C for up to 3 months.
Ampicillin stock solution (50 mg/ml)
Dissolve 0.5 g of ampicillin sodium salt in 10 ml of distilled H2O. Sterilize the solution by filtering with a 0.22 µm Ultrafree-CL PVDF filter column. Store stock solution at −20 °C for up to 5 years.
Agarose gel, 1%/2% (wt/vol)
Add 1 g (1%) or 2 g (2%) of agarose to 100 ml of TAE buffer and boil until the agarose is completely dissolved. Let it cool down to ~50 °C. Add 5 µl of SYBR Safe DNA Gel Stain per 100 ml gel (1:20,000 dilution), and pour the gel in a casting chamber. Gels can be stored at 4 °C for up to 7 d.
Lysogeny broth medium (LB medium)
Dissolve 10 g of tryptone, 5 g of yeast extract and 5 g of sodium chloride in 1 L of distilled water. Autoclave and store the medium at 4–8 °C for up to 3 years.
LB-ampicillin agar plates
Dissolve 15 g of agar bacteriology grade in 1 L of LB medium and sterilize by autoclaving. Let the solution cool down to room temperature and add 1 ml of ampicillin stock solution. Pour the solution in Petri dishes and let it cool down with lids slightly open. Store plates at 4 °C for up to 1 month.
preTB medium
Dissolve 12 g of tryptone, 24 g of yeast extract and 5 ml of glycerol in 1 L of distilled water. Autoclave the solution and store at 4 °C for up to 3 years.
TB salt buffer (10×)
Dissolve 125.4 g of K2HPO4 and 23.1 g of KH2PO4 in 1 L of distilled water. Autoclave the solution and store at 4 °C for up to 5 years.
Terrific broth medium (TB medium)
Add 100 ml of TB salt buffer (10×) to 900 ml of preTB medium. Store TB medium at 4 °C for up to 3 years.
PEI, 0.045% (wt/vol)
Dilute 0.45 g of PEI in 1 L of ddH2O and dissolve under constant stirring. Filter solution through a 0.22 µm membrane filter. Store PEI solution for 1 year at room temperature.
Primers
Dilute primers with nuclease-free water to a stock concentration of 50 µM. For primer mixes, add the same volume of each primer (Supplementary Tables 1–9). Store primers and primer mixes at −20 °C for up to 3 years.
HEK293-6E medium
Add 2 ml of penicillin–streptomycin (10,000 U/ml) to 1 L of FreeStyle 293 expression medium. Store medium at 4–8 °C for up to 3 months.
HEK293T complete medium
Add 50 ml of FBS, 5 ml of antibiotic–antimykotic, 5 ml of sodium pyruvate and 5 ml of l-glutamine to 435 ml of DMEM high-glucose medium. Store medium at 4–8 °C for up to 3 months.
HEK293T starving medium
Add 10 ml of FBS, 5 ml of antibiotic–antimykotic, 5 ml of sodium pyruvate and 5 ml of l-glutamine to 475 ml of DMEM high-glucose medium. Store medium at 4–8 °C for up to 3 months.
Tris-HCL buffer, 1 M, pH 8.0
Dissolve 12.1 g of Tris-(hydroxymethyl)-aminomethane in 80 ml of distilled H2O. Adjust pH to 8.0 with hydrochloric acid. Fill up the final volume to 100 ml with distilled H2O. Autoclave and store at room temperature for up to 5 years.
Glycine 0.1 M, pH 3.0
Dissolve 0.75 g of glycine in 80 ml of distilled H2O. Adjust the pH to 3.0 with hydrochloric acid. Fill up the final volume to 100 ml with distilled H2O. Autoclave and store at room temperature for up to 5 years.
ELISA washing buffer
0.05% Tween-20 in 1× PBS. Store buffer at room temperature for up to 5 years.
Blocking solution
2% BSA and 0.1% Tween-20 in 1× PBS. Store blocking solution at 4–8 °C for a maximum of 2 d.
Procedure
Stage I: antigen-specific B-cell isolation
Timing 2.5 h (day 1)
PBMC collection
Caution
Sample collections must be performed in accordance with the study protocols approved by the local IRB. Informed consent must be obtained from all study participants.
-
1
Obtain starting material for the isolation of PBMCs.
Critical step
PBMCs can be obtained from numerous sources, including peripheral blood, leukapheresis or buffy coats. Note that B cells can also be isolated from tissues such as tonsils or lymph nodes. Different sources for human sample collection determine the overall yield of cells and might therefore limit the downstream investigation of B cells.
Pause point
Fresh blood samples with an appropriate anticoagulant can be stored at room temperature under constant shaking for up to 16 h prior to PBMC isolation. Longer incubation periods will reduce the number of viable cells.
-
2
Perform PBMC isolation by standard density gradient centrifugation using Histopaque separation medium and Leucosep cell tubes according to the manufacturer’s protocol.
Pause point
Cells can be cryopreserved for over 10 years at −150 °C.
Critical step
The use of appropriate freezing and thawing protocols is critical to ensure recovery of viable cells.
-
3
If cryopreserved PBMCs are obtained, thaw cells quickly for 1 min in a water bath at 37 °C and transfer cells immediately into a 50 ml tube containing cold 1× PBS (4–8 °C).
Isolation of CD19+ B cells
Timing 1.5 h (day 1)
Critical step
Keep cell suspension cool and use cold reagents (4 °C) to prevent nonspecific binding of CD19 microbeads to cells.
-
4
Proceed with freshly isolated or thawed cells from Step 3.
-
5
Centrifuge cell suspension at 4 °C and 400g for 10 min.
-
6
Carefully discard supernatant and resuspend cell pellet in 80 µl of MACS buffer per 107 cells (for preparation of MACS buffer, see ‘Reagent setup’).
-
7
Add 20 µl of CD19 microbeads per 107 cells to the suspension.
-
8
Mix well and incubate at 2–8 °C in a refrigerator for 15 min.
Critical step
Longer incubation times and higher temperatures might lead to unspecific binding of CD19 beads.
-
9
Add 10 ml of cold MACS buffer (4–8 °C) to the cell suspension and centrifuge for 10 min at 400g and 4 °C. Discard supernatant.
-
10
Resuspend the cell pellet in 0.5 ml of MACS buffer.
-
11
Place LS columns in the magnetic field of the magnetic separator, place a collection tube (15 ml plastic tube) below each column, equilibrate each column with 3 ml of MACS buffer and let the columns drain by gravity.
-
12
Apply cell suspension to equilibrated LS columns and let the columns drain by gravity into the collection tube.
Critical step
It is recommended to pass the cell suspension through a preseparation filter of 70 µm to prevent the LS columns from clogging.
-
13
Wash the cell suspension tube with an additional 500 µl of MACS buffer and apply to LS columns.
-
14
Wash the LS column three times with 3 ml of MACS buffer.
-
15
Remove the LS column from the magnetic separator and place it on a new 15 ml collection tube.
-
16
Add 5 ml of MACS buffer onto the column and immediately flush out CD19+ cells bound to magnetic beads by firmly pushing the plunger into the column.
-
17
Determine cell number and centrifuge cell suspension for 10 min at 400g and 4 °C.
Staining of antigen-specific B cells and single-cell sorting
Timing 3 h (day 1)
Critical step
For antigen labeling, we successfully used recombinant fusion to GFP or chemical coupling by either NHS-ester chemistry (DyLight labeling kits according to the manufacturer’s manual) or AviTag-mediated biotinylation (BirA500 kit) followed by PE-streptavidin staining as described elsewhere35.
-
18
Resuspend cell pellet in 100 µl of FACS buffer per 1 × 106 isolated B cells in a 15 ml plastic tube and keep cells on ice.
-
19
Prepare master mix of fluorescently labeled antibodies and fluorochrome-conjugated antigen(s) and add to cells on ice.
Critical step
Antigen-specific B cells should be distinguishable from nonspecific B cells. Optimization of staining with labeled antibodies and conjugated antigen(s) by titration and the use of negative controls is essential.
Dilutions of antibodies and staining reagents used for single-cell sorts
Antibody/stain
Dilution
Final concentration
FITC mouse anti-human IgM
1:5
na
Alexa Fluor 700 mouse anti-human CD20
1:80
na
APC mouse anti-human IgG
1:20
na
DAPI (300 µM stock solution)
1:100
3 µM
-
20
Incubate antibodies and antigens for 20 min on ice and in the dark.
Dilutions of fluorescently labeled antigens used for single-cell sorts
Labeled antigena
Final concentration
Dye
Conjugation method
Estimated dye/protein ratio
BG505 SOSIP.664 gp140
15 µg/ml
GFP
Fusion protein
1:1
YU2gp140
15 µg/ml
PE
Biotin–Streptavidin
na
EBOV GP∆TM
10 µg/ml
DyLight 488
NHS Ester
4:1
SARS-CoV-2 spike
10 µg/ml
DyLight 488, DyLight 650
NHS Ester
2:1–8:1
-
21
In the meantime, prepare sorting buffer and add 4 µl of buffer to each well of a 96-well PCR plate:
Composition of sorting buffer
Component
Amount per well (µl)
Final concentration
Nuclease-free H2O
3.1
RNasin (40 U/µl)
0.2
2 U/µl
RNaseOut (40 UL)
0.1
1 UL
PBS (10×)
0.2
0.05×
DTT (100 mM)
0.4
1 mM
Sum
4
-
22
Centrifuge 96-well PCR plate for 30 s at 800g and 4 °C.
-
23
Store 96-well PCR plates at 4–8 °C in the refrigerator until single-cell sort.
Critical step
Label plates with date, patient ID, fluorochrome-conjugated antigen and plate number. Maintain this labeling system throughout the following steps of the protocol.
-
24
For washing after the staining in Step 20, fill up the plastic tube to 15 ml with FACS buffer and centrifuge for 10 min at 400g and 4 °C.
-
25
Discard supernatant and resuspend cell pellet in 100 µl FACS buffer per 106 cells (for preparation of FACS buffer, see ‘Reagent setup’). Samples with less than 3.5 × 106 cells are resuspended in 350 µl FACS buffer.
-
26
Transfer cell suspension through a cell strainer cap into a 5 ml polystyrene round-bottom tube.
-
27
Proceed with compensation setup and gating on the single-cell sorter.
-
28
Adjust gates: identify lymphocyte population in FCS-A/SSC-A and single cells in FCS-H/FCS-A. Display cells in FCS-A/DAPI and gate into DAPI negative cells. To identify B cells, plot FCS-A/CD20 cells and gate into CD20+ cells. Continue with a plot for CD20/IgG cells and gate into CD20+ and IgG+ cells. To set up the final sorting gate, display the bait protein against IgG and gate to the bait+ and IgG+ fraction. In case of double staining with two differently labeled baits, gate on IgG+ cells in the FCS-A/IgG plot first; then display the two colors of the differently labeled bait proteins and place a sorting gate on double positive cells.
Critical step
Be precise with placement of the gates to avoid unspecific cell sorting.
Critical step
To identify the bait-positive population, it is important to include negative and positive controls, if possible.
-
29
Proceed with single-cell sorts into 96-well PCR plate prefilled with sorting buffer. Spare the last 3–12 wells as negative controls for following protocol steps.
Critical step
Spare wells are used as negative controls for subsequent single-cell PCRs to detect nucleic acid contaminations.
-
30
After sorting, immediately freeze and store 96-well sorting plates at −80 °C.
Stage II: high-efficiency BCR amplification
Reverse transcription
Timing 2 h (day 1)
Critical step
It is critical to work carefully and to prevent any contamination. Use sterile filter pipette tips and a designated DNA/RNase-free hood to prepare master mixes and plates for reverse transcription and single-cell PCR reactions. The hood and all surfaces should be cleaned with DNA/RNase AWAY prior to working. Wipe all reagent tubes with a DNA/RNase AWAY treated tissue before putting them into the PCR hood. We recommend to only prepare master mixes or handle cells and RNA samples under the PCR hood. Pipetting of cDNA or DNA templates should be performed outside of the hood on a dedicated workbench.
-
31
Thaw reagents on ice.
-
32
First, prepare random-hexamer-primer (RHP) master mix under the hood and place it on ice.
Composition of RHP master mix
Component
Amount per well (µl)
Amount per plate (µl)
Final concentration
Nuclease-free H2O
5.6
560
Random hexamer primers (200 ng/µl)
0.75
75
21 ng/µl
NP40 (10% (vol/vol))
0.5
50
0.71% (vol/vol)
RNaseOut (40 U/µl)
0.15
15
0.9 U/µl
Sum
7
700
-
33
Prepare reverse-transcription (RT) master mix without SuperScript IV reverse transcriptase under the hood and place it on ice.
Critical step
SuperScript IV is added at Step 41 after primer annealing.
Composition of RT master mix
Component
Amount per well (µl)
Amount per plate (µl)
Final concentration
Nuclease-free H2O
2.05
205
SuperScript IV RT Buffer (5×)
3
300
2.1×
dNTP mix (25 mM)
0.5
50
1.8 mM
DTT (100 mM)
1
100
14 mM
RNasin (40 U/µl)
0.1
11
0.57 U/µl
RNaseOut (40 U/µl)
0.1
11
0.57 U/µl
Intermediate sum
6.75
67.5
SuperScript IV (200 U/µl)
0.25
25
7.14 U/µl
Final sum
7
700
-
34
Thaw 96-well sorting plates from single-cell sorts on ice and centrifuge briefly to collect the content on the bottom of each well.
-
35
Under the hood, transfer RHP master mix in a pipetting reservoir.
-
36
Add 7 µl of RHP mix with a multichannel pipette to each well of the 96-well sorting plate.
-
37
Homogenize the sample by pipetting up and down 20× and rinse the walls of each well twice to assure the capturing of cells that were not sorted into the center of the well.
-
38
Homogenize again by pipetting up and down 10× and seal the plate with adhesive foil.
-
39
Incubate the 96-well sorting plate in a thermocycler at 65 °C for 2.5 min.
-
40
Place the plate on ice for at least 2 min.
-
41
Add SuperScript IV to RT master mix.
-
42
Transfer RT master mix in a pipetting reservoir, add 7 µl of RT master mix with a multichannel pipette to each well of the 96-well sorting plate and resuspend by pipetting up and down 3×.
-
43
Centrifuge the plate at 800g.
-
44
Place the plate in a thermocycler and run the reverse transcription program.
Cycle parameters of reverse transcription
Temperature (°C)
Duration (min)
42
10
25
10
50
10
94
5
4
∞
-
45
Dilute the cDNA with 16 µl of nuclease-free water.
-
46
Add ‘cDNA’ to the label of the 96-well sorting plate.
-
47
Store plate at −80 °C, or proceed to first PCR.
Pause point
cDNA can be stored for up to 10 years at −80 °C.
First single-cell PCR for amplification of heavy- and light-chain genes
Timing 3 h (day 1)
-
48
Proceed with diluted cDNA from Step 45, or thaw plates on ice.
-
49
Clean the hood with DNA/RNase AWAY and use filtered pipette tips.
-
50
Thaw reagents on ice.
-
51
Heavy- and light-chain variable gene regions are preamplified in a single PCR.
Critical step
For amplification of variable gene regions with high coverage, optimized multiplex primer sets are used. We also provide primers for IgM heavy-chain amplification, if IgM+ cells have been sorted (see Supplementary Tables 1 and 3, and refer to ‘Reagent setup’ for preparation of first single-cell PCR primer mixes).
-
52
Prepare the first PCR master mix under the hood.
Composition of first single-cell PCR master mix
Component
Amount per well (µl)
Final concentration
Nuclease-free H2O
14.68
Platinum Taq PCR buffer (10×)
2.05
0.9×
KB Extender (6%)
1.23
MgCl2 (50 mM)
0.61
1.34 mM
dNTP mix (25 mM)
0.16
0.18 mM
oPR_1st_fwd primer mix (50 µM)
0.09
0.2 µM
oPR_1st_IgG_rev or oPR_1st_IgM_rev primer mix (50 µM)
0.09
0.2 µM
Platinum Taq DNA Polymerase
0.09
Sum
19.0
-
53
Use a multichannel pipette to transfer 19 µl of first PCR master mix to each well of a new 96-well PCR plate.
Critical step
For each cDNA plate, you will produce one first PCR plate. Label the 96-well PCR plate appropriately with date, patient ID, fluorochrome-conjugated antigen, plate number and ‘first PCR’.
-
54
Outside of the hood, use a multichannel pipette to add 3.8 µl from each well of the cDNA plate to the same well of the first PCR plate.
-
55
Seal the plate with adhesive foil and run the first single-cell PCR thermocycler program.
Cycle parameters of first single-cell PCR
Temperature (°C)
Duration
No. of cycles
94
2 min
1
94
30 s
50
57
30 s
50
72
55 s
50
72
6 min
1
4
∞
1
-
56
Store first PCR plates at −20 °C.
Pause point
First PCR products can be stored for up to 3 years at −20 °C.
Second single-cell PCR for amplification of heavy- and light-chain genes
Timing 3 h (day 2)
-
57
Proceed with first PCR plates from Step 55, or thaw first PCR plates on ice.
-
58
Clean hood with DNA/RNase AWAY and work with filtered pipette tips.
-
59
Thaw reagents on ice.
-
60
Targeted amplification of heavy- and the two light-chain variable regions is performed in three separate reactions.
-
61
Prepare the second PCR master mixes for each chain under the hood.
Composition of second single-cell PCR master mix
Component
Amount per well (µl)
Final concentration
Nuclease-free H2O
14.68
Platinum Taq Green PCR buffer (10×)
2.05
0.9×
KB Extender (6%)
1.23
MgCl2 (50 mM)
0.61
1.4 mM
dNTP mix (25 mM)
0.16
0.18 mM
5′ primer mix (50 µM)
0.09
0.2 µM
3′ primer (50 µM)
0.09
0.2 µM
Platinum Taq DNA polymerase
0.09
Sum
19.0
-
62
Use a multichannel pipette to transfer 19 µl of second PCR master mix to each well of a 96-well PCR plate.
Critical step
For each first PCR plate, you will produce three second PCR plates. Label the 96-well PCR plates appropriately with date, patient ID, fluorochrome-conjugated antigen, plate number, antibody chain (heavy, kappa and lambda chain) and ‘second PCR’.
-
63
Outside of the hood, use a multichannel pipette to transfer 3 µl of the first PCR product from each well to the same well of the second PCR plate. Repeat this step for all three chains (heavy, kappa and lambda).
-
64
Seal the plates with adhesive foil and run the following thermocycler program.
Cycle parameters of second single-cell PCR
Temperature (°C)
Duration
No. of cycles
94
2 min
1
94
30 s
50
57
30 s
50
72
45 s
50
72
6 min
1
4
∞
1
-
65
Prepare a 2% agarose gel according to ‘Reagent setup’.
-
66
Load 6 µl of 1 kb plus ladder (0.5 µg) and 3 µl of second PCR product per lane.
-
67
Let the gel run at 120 V for 30 min in 1× TAE buffer and evaluate which chains have been amplified.
Critical step
Expected product lengths for heavy chains will be around 500 bp and 450 bp for light chains.
-
68
Send in amplicons (~19 µl PCR product will be left after gel electrophoresis) for sequencing with the respective chain-specific reverse primer that was used for the second PCR.
Critical step
The reverse primer of the second PCR anneals to the constant region and allows sequencing of the complete V(D)J region.
-
69
Second PCR plates can be stored overnight at 4 °C, if sealed properly.
Pause point
For long-term storage, second PCR product can be stored at −20 °C for up to 3 years.
Sequence annotation, analysis and selection for downstream antibody production
Timing 2–4 h (day 3)
-
70
Investigate second PCR sequences for their overall quality (by phred score or manually) and propagate high-quality sequencing results only (we allow up to 15 nucleotides within the variable region to have phred scores below 16).
-
71
Use annotation software such as IgBLAST76 or V-QUEST79 to annotate heavy- and light-chain sequences.
-
72
Prepare an overview of all analyzed B cells where you enter a unique plate name, the well, and heavy- and light-chain sequence information including their quality, V(D)J gene information, V gene germline identity, CDR3 sequence in amino acids, and the trimmed V region nucleotide sequence.
Critical step
Either continue with all cells or prepare a selection of antibodies that should be cloned and transfer first single-cell PCR products from Step 56 of selected cells to a fresh multiwell PCR plate. If cells are selected, transferred first PCR products can be sorted by chain type to allow convenient pipetting of third PCR master mixes. Carefully document which sequences have been moved to which position on the new compilation PCR plate. We typically select sequences for downstream antibody cloning and production according to sequence features and clonal relationships. For further information on criteria for sequence selection, refer to Experimental design, section High efficiency BCR amplification (II).
Stage III: high-efficiency BCR cloning
Cloning PCR of heavy- and light-chain genes
Timing 2 h (day 3)
Critical step
SLIC requires homologous regions between vector backbones and inserts. We have established optimized multiplexed primer sets for the amplification of all heavy- and light-chain V genes from first PCR products for the subsequent cloning into mammalian expression vectors. For detailed information about SLIC primer sets, see Supplementary Tables 2, 4 and 8.
-
73
Proceed with first PCR plate from Step 56.
-
74
Thaw first PCR plate on ice.
-
75
Clean the hood with DNA/RNase AWAY and work with filtered pipette tips.
-
76
Thaw reagents on ice.
-
77
Prepare the appropriate number of heavy- and light-chain cloning PCR master mixes under the hood.
Critical step
Composition of cloning PCR master mix
Component
Amount per well (µl)
Final concentration
Nuclease-free H2O
24.1
Q5 PCR buffer (5×)
10
1×
dNTP mix (25 mM)
0.4
0.21 mM
5′ primer mix (50 µM)
0.5
0.5 µM
3′ primer (50 µM)
0.5
0.5 µM
Q5 High GC Enhancer (5×)
10
1×
Q5 high-fidelity DNA polymerase
0.5
Sum
46
-
78
Add 46 µl of cloning PCR master mix to each well of a 96-well PCR plate.
Critical step
For each first PCR plate, you will produce two third PCR plates (one for the heavy and one for the light chain). Label the 96-well PCR plates appropriately with date, patient ID, fluorochrome-conjugated antigen, plate number, antibody chain and ‘third PCR’.
-
79
Outside the hood, use a multichannel pipette to transfer 1 µl of the first PCR product from each well to the same well of the heavy-chain cloning PCR plate. Repeat this step for the light-chain cloning PCR plate.
Critical step
Do not add more than 1 µl of first PCR product, as this will result in unspecific amplification. Unspecific products will inhibit subsequent cloning reaction.
-
80
Seal the plate with adhesive foil and run the two-step cloning PCR program on a thermocycler.
Cycle parameters for cloning PCR
Temperature (°C)
Duration
No. of cycles
98
30 s
1
98
10 s
35
72
45 s
35
72
2 min
1
4
∞
1
-
81
Prepare a 2% agarose gel according to ‘Reagent setup’.
-
82
Load 6 µl of 1 kb plus ladder (0.5 µg) and 4 µl of cloning PCR product per lane.
-
83
Let the gel run at 120 V for 30 min in 1× TAE buffer and analyze the gel for amplified product.
Critical step
Expected product lengths for heavy chains will be around 600 bp and 550 bp for light chains.
Pause point
Cloning PCR product can be stored at −20 °C for up to 3 years.
Linearization of expression vector
Timing 8–16 h (days 2–3)
Critical step
-
84
Thaw IgG1, IgK and IgL vectors at room temperature.
-
85
Prepare the restriction digests for the respective vectors in separate 1.5 ml safe-lock tubes.
Critical step
Always handle restriction enzymes on ice or in a benchtop cooler. Be careful to avoid cross-contamination of different restriction enzymes or enzymes and expression vectors. For restriction digest, we recommend at least a tenfold overdigestion of the backbones to prevent transformation of undigested vector. Total enzyme volume should not exceed 10% of the whole mix volume to prevent star activity.
Composition of restriction digests
IgG1 (heavy chain)
IgK (kappa chain)
IgL (lambda chain)
10 µg vector
10 µg vector
10 µg vector
2.5 µl EcoRI-HF (50 U)
2.5 µl EcoRI-HF (50 U)
2.5 µl EcoRI-HF (50 U)
2.5 µl SalI-HF (50 U)
2.5 µl BsiWI-HF (50 U)
2.5 µl XhoI (50 U)
5 µl CutSmart Buffer
5 µl CutSmart Buffer
5 µl CutSmart Buffer
40 µl nuclease-free H2O
30 µl nuclease-free H2O
30 µl nuclease-free H2O
-
86
Incubate the reaction for 2–16 h at 37 °C.
Critical step
Incubation times of <2 h might lower cloning efficiency as the vector will not be restricted completely. Recommended incubation time is 16 h (overnight). In the meantime, proceed with PCR product purification of cloning PCR from Step 80.
-
87
For denaturation of restriction enzymes, incubate the reaction at 65 °C for 20 min.
-
88
Add 0.5 µl of CIP and incubate at 37 °C for 30 min.
-
89
Prepare a 1% agarose gel according to ‘Reagent setup’.
-
90
Load 6 µl of 1 kb plus ladder (0.5 µg) and the whole volume of each restriction master mix into separate gel pockets.
-
91
Let the gel run at 120 V for 30 min to 1 h in 1× TAE buffer.
-
92
Expose the gel to UV light and excise restricted vectors with a scalpel.
Critical step
Use a new scalpel for different vectors to prevent cross-contamination. Exposure time to UV light should be as short as possible, as UV light damages DNA.
-
93
Proceed with gel purification of restricted vectors using NucleoSpin Gel and PCR Clean-up Kit.
-
94
Adjust concentration of restricted vectors to 80 ng/µl with nuclease-free water.
Pause point
Restricted vectors can be stored at −20 °C for up to 3 years.
SLIC of antibody variable regions
Timing 4 h (day 3)
-
95
Thaw cloning PCR plates from Step 80 on ice and purify PCR product with the NucleoSpin 96 PCR Clean-up Kit.
Critical step
Purification of cloning PCR plates can be conducted during vector restriction (see Step 86).
-
96
Adjust concentration of purified DNA to 20–40 ng/µl with nuclease-free water.
Critical step
A molar ratio of PCR product to vector of 2:1–6:1 is optimal for SLIC.
-
97
Set thermocyclers to 24 °C for the cloning reaction.
-
98
Thaw chemically competent E. coli DH5α cells on ice.
-
99
Prepare SLIC master mix in a safe-lock tube or conical plastic tube on ice.
Composition of SLIC master mix
Component
Amount per well (µl)
Final concentration
Nuclease-free H2O
6.8
Linearized expression vector (80 ng/µl)
1
8 ng/µl
NEBuffer 2.1 (10×)
1
1×
T4 DNA polymerase (3,000 U/ml)
0.2
60 U/ml
Sum
9
Critical step
T4 DNA polymerase has 3′→5′ exonuclease activity. Prepare and handle SLIC master mix strictly on ice to avoid uncontrolled digestion of DNA. Start the reaction by incubation at 24 °C in a thermocycler.
-
100
Label a 96-well PCR plate with SLIC-MM and the respective plate name. Keep all plates on ice.
-
101
Transfer 9 µl of SLIC master mix to the SLIC-MM plate with a multichannel pipette (on ice).
-
102
Transfer 1 µl of purified cloning PCR product of each well to the same well on the SLIC-MM plate (on ice).
-
103
Seal the SLIC-MM plate with foil and incubate at 24 °C for exactly 2.5 min in a thermocycler.
Critical step
Longer or shorter incubation times might lower the efficiency of the cloning reaction.
-
104
Immediately incubate the SLIC-MM plate on ice for at least 10 min.
-
105
Set the thermocycler to 42 °C.
-
106
Add 40 µl of chemically competent DH5α cells to each well of a new 96-well PCR plate and label the plate with transformation-MM.
-
107
Transfer 4 µl of SLIC reaction from the SLIC-MM to the same wells of the transformation-MM plate with a multichannel pipette.
-
108
Incubate the transformation-MM plate on ice for at least 30 min.
Critical step
Shorter incubation times will decrease the transformation efficiency.
-
109
Provide the appropriate number of LB-ampicillin agar plates (see ‘Reagent setup’) at room temperature. Each well of a transformation-MM plate will be transferred to a corresponding agar-plate; i.e., 96 agar plates will be needed per 96-well plate.
-
110
Incubate the transformation-MM plate in a thermocycler at 42 °C for 45 s.
Critical step
The heat shock will enhance the efficiency of the transformation and is strongly recommended. However, it is not mandatory for vector uptake in chemically competent cells.
-
111
Incubate the transformation-MM plate on ice for 2 min.
-
112
Add 50 µl of SOC medium to each well of the transformation-MM plate using a multichannel pipette.
-
113
Tightly seal the plate with adhesive foil and incubate the plate on a bacterial shaker at 37 °C and 210 rpm for 1 h.
-
114
In the meantime, label agar plates with plate identifier, well number and chain information.
Critical step
For a convenient labeling of large amounts of agar plates, we recommended printable heavy-duty labels.
-
115
After incubation (Step 113), briefly centrifuge the transformation-MM plates at a maximum of 200g for 10 s to collect evaporated medium from the foil.
-
116
Streak the whole volume of each well on the corresponding labeled LB-ampicillin agar plate.
-
117
Incubate agar plates at 37 °C for 16 h.
Pause point
Agar plates can be sealed with Parafilm and stored at 4 °C for up to 4 weeks.
Colony PCR and sequencing
Timing 24 h (day 4)
-
118
Provide agar plates with transformed colonies from Step 117.
-
119
For each agar plate, three colonies are examined by colony PCR for correct insertion of the variable region into the expression vector. Each colony will also be used to inoculate a liquid culture. For this purpose, prepare three 96-well PCR plates with colony PCR master mixes and three Microtest 96-well plates with LB medium with ampicillin.
-
120
Prepare the colony PCR master mix on ice.
Composition of colony PCR master mix
Component
Amount per well (µl)
Final concentration
Nuclease-free H2O
17.44
Platinum Taq Green PCR buffer (10×)
2.20
1×
KB Extender (6%)
1.28
MgCl2 (50 mM)
0.65
1.6 mM
dNTP mix (25 mM)
0.2
0.23 mM
5′ Ab-sense (50 µM)
0.08
0.18 µM
3′ primer (50 µM)
0.08
0.18 µM
Platinum Taq DNA polymerase
0.08
Sum
22
-
121
Add 22 µl of colony PCR master mix to each well of three 96-well PCR plates using a multichannel pipette.
Critical step
Label each plate appropriately with date, patient ID, fluorochrome conjugated bait, plate number, antibody chain, and cPCR I/II or III.
-
122
Dilute ampicillin stock solution in prewarmed LB medium to reach a recommended working concentration of 50 µg/ml.
-
123
Prefill each well of three Microtest 96-well plates with 50 µl of the LB-ampicillin solution.
Critical step
Label each liquid culture plate appropriately with date, patient ID, plate number, antibody chain, and cPCR I/II or III.
-
124
With a sterile pipette tip, pick a single bacterial colony from the first labeled LB-ampicillin agar plate, i.e., the transformation corresponding to the third PCR product in well A1.
-
125
Inoculate the respective well A1 of the prefilled liquid culture plate cPCR I with the first colony.
Critical step
Be careful not to cross-contaminate the wells of the liquid culture plates.
-
126
After inoculation, transfer the same tip to the colony PCR plate cPCR I in the corresponding well A1.
-
127
Repeat these steps with a second and a third colony from the same agar plate: inoculate the corresponding well A1 of the liquid culture plates cPCR II and III with the respective colony before transferring the tips to A1 of the colony PCR plates II and III, respectively.
-
128
Proceed with the next agar plate (i.e., the transformation corresponding to third PCR product in well number A2), pick three colonies to inoculate the three liquid culture plates cPCR I, II and III in well A2 and transfer the tips to A2 of the respective colony PCR plates I, II and III. Repeat these steps for all agar plates.
-
129
Incubate liquid culture plates cPCR I, II and III at 37 °C for 16 h.
-
130
Run the colony PCR on a thermocycler with the colony PCR program.
Cycle parameters for colony PCR
Temperature (°C)
Duration
No. of cycles
94
5 min
1
94
30 s
28
55
30 s
28
72
1 min
28
72
5 min
1
4
∞
1
-
131
Prepare a 2% agarose gel according to ‘Reagent setup’.
-
132
Load 6 µl of 1 kb plus ladder (0.5 µg) and 4 µl of colony PCR product per lane.
-
133
Run the gel at 120 V for 30 min in 1× TAE buffer.
Critical step
Expected product lengths will be around 680 bp for heavy and 630 bp for light chains.
-
134
Send DNA of wells with correct product lengths for sequencing with 5′ Ab-sense primer.
Pause point
LB-ampicillin plates can be stored at 4 °C for up to 4 weeks. Colony PCR plates can be stored at −20 °C for up to 3 years.
Stage IV: high-throughput and/or large-scale antibody production
-
135
Conduct either high-throughput (option A) or large-scale (option B) antibody production.
-
(A)
High-throughput antibody production
96-well mini DNA preparation (high-throughput)
Timing 48 h (days 5–7)
-
(i)
Align sequences of colony PCR products to sequences from second PCR products (Step 68).
-
(ii)
Identify correct cPCR-clones for each heavy and light chain. To account for PCR and sequencing errors, we allow for a difference of up to three nonsynonymous mutations between second PCR and colony PCR products, as long as they do not occur in the complementarity-determining regions (CDRs).
-
(iii)
Either combine liquid cultures of correct clones on a new plate with the same layout or continue inoculation of bacterial cultures directly from the three individual cPCR liquid cultures.
Critical step
After inoculation of bacterial cultures for plasmid-DNA preparation, cPCR liquid cultures can directly be stored as glycerol stocks by adding 50 µl sterile glycerol to the 50 µl liquid culture, mixing, sealing and storing the plate at −80 °C.
-
(iv)
For inoculation and subsequent mini-scale plasmid-DNA preparations, use the NucleoSpin 96 Plasmid kit.
-
(v)
Clean workspace and pipettes with 70% ethanol and DNA/RNase AWAY.
-
(vi)
Add 150 ml of TB medium per 96-well plate (see ‘Reagent setup’) at room temperature at 37 °C and 150 µl of the ampicillin stock solution to a final concentration of 50 µg/ml.
-
(vii)
Use a multistep pipette to add 1.5 ml of TB-ampicillin medium to each well of a 96-square-well culture plate enclosed in the NucleoSpin 96 Plasmid kit.
Critical step
Do not contaminate the pipette tip or spill medium between wells.
-
(viii)
Inoculate each well either directly from the liquid culture or respective 96 glycerol stock plate cPCR I/II or III from Step 135A(iii).
-
(ix)
Seal the square-well culture plate with the enclosed woven cover membrane.
-
(x)
Incubate the square-well culture plate in a bacterial shaker at 37 °C and 230 rpm for 18–36 h.
Critical step
Do not incubate cultures longer than 48 h as this will lead to a higher percentage of dead bacterial cells.
-
(xi)
Centrifuge square-well culture plates for 10 min at room temperature and 1,000g.
Critical step
Exceeding 1,000g will lead to tight clogging of bacterial pellets and will complicate resuspension in the following steps.
-
(xii)
Discard supernatant by gently flipping the square-well plates upside down and tapping on paper towels.
Critical step
To prevent cross contaminations, do not flip the plates back to upside before tapping residual fluid on paper towels.
-
(xiii)
Follow the enclosed manual of the NucleoSpin 96 Plasmid kit for mini-scale plasmid DNA preparation.
Critical step
We expect DNA concentrations of 500–1,000 ng/µl and yields of 20–50 µg, which typically cover 10–20 transfections in a 48-well format or 20–40 transfections in a 96-well format.
-
(xiv)
Sequence mini-scale plasmid DNA preparations with the 5′ Ab-sense primer.
Pause point
Mini-scale plasmid-DNA preparations can be stored at −20 °C for up to 3 years.
Transfection of HEK293T cells (high-throughput)
Timing 3 h (days 8–12)
-
(xv)
Align V region sequences from 96-well mini-scale plasmid DNA preparations to the respective second PCR sequence from Step 68 and the cPCR product from Step 134 to make sure to propagate the correct plasmid.
Critical step
This is the last possibility to check that correct antibody heavy- and light-chain pairs will be transfected.
-
(xvi)
Transfection of antibody plasmids can be conducted in either a 48- or 96-well format in cell culture plates. Prepare the following transfection mixes:
Cell culture and transfection volumes for high-throughput antibody production
Plate format
Seeding volume (µl/well)
Number of cells/well
Amount of heavy- and light-chain plasmid (µg each/well)
Volume of diluted plasmid DNA (µl/well)
Total volume of transfection master mix (µl/well)
Volume of transfection reagent (µl/well)
Final volume for transfection (µl/well)
48-well plate
400
6 × 104
0.2
20
20
1
40
96-well plate
200
4 × 104
0.1
10
10
0.4
20
-
(xvii)
Working under a sterile hood with laminar flow, seed HEK293T cells per well according to the chosen cell culture plate format in starving medium (see ‘Reagent setup’).
Critical step
Higher cell numbers might lower transfection efficiency. Use starving medium for seeding to prevent cell culture from overgrowth.
-
(xviii)
Incubate cell culture plates at 37 °C for 18–24 h in a cell culture incubator.
-
(xix)
Adjust the concentrations of heavy- and light-chain plasmids to 50 ng/µl with nuclease-free water.
-
(xx)
Transfer the appropriate amount of heavy and corresponding light-chain plasmid DNA in each well of a new 96-well PCR plate (plasmid transfection plate). Fill up each well with serum-free DMEM to a final volume of 20 µl (48-well) or 10 µl (96-well), respectively.
Critical step
Use one well as a control well for transfection efficiency by transfecting the same total amount of GFP-expressing plasmid.
-
(xxi)
Calculate the total volume of Turbofect transfection reagent and serum-free DMEM needed for transfection master mix. For example, for transfection of a single 48-well cell culture plate, 48 µl of transfection reagent is diluted in 912 µl of serum-free DMEM to reach a final volume of 960 µl of transfection master mix.
-
(xxii)
In a 1.5 ml safe-lock tube, prepare the transfection master mix and briefly vortex for 30 s.
-
(xxiii)
Add the appropriate volume of transfection master mix to each well of the plasmid transfection plate containing diluted antibody plasmids.
-
(xxiv)
Seal the transfection plate tightly with plastic flap cap strips and vortex the plate for 1 min.
Critical step
For the formation of transfection complexes, it is important to vortex thoroughly.
-
(xxv)
Incubate transfection plate at room temperature for 15–20 min.
-
(xxvi)
Add 40 or 20 µl of each well of the transfection plate dropwise to the corresponding well of the 48- or 96-well cell culture plate from Step 135A(xvii).
Critical step
It is important to gently rock cell culture plates to ensure even distribution of the transfection complexes within each well.
-
(xxvii)
Incubate the transfected cell culture plate for 96 h at 37 °C in a cell culture incubator.
-
(xxviii)
24 h after transfection, check control wells by fluorescence microscopy for GFP expression.
-
(xxix)
96 h after transfection, centrifuge the cell culture plate at 400g and room temperature for 5 min.
-
(xxx)
Transfer supernatants to a new Microtest 48- or 96-well plate.
Pause point
Supernatants can be stored for 10 d at 4 °C.
Human IgG capture ELISA (high-throughput)
Timing 6 h (day 12)
Critical step
Use a multichannel pipette and ELISA washer for the following steps: xxxii, xxxv, xxxvi, xxxviii–xl, xliii, xliv, xlvii, xlviii.
-
(xxxi)
Adjust concentration of polyclonal goat-anti-human IgG to 2.5 µg/ml in 1× PBS.
-
(xxxii)
Coat each well of a 96-well high-binding ELISA plate with 50 µl of polyclonal goat-anti-human IgG.
-
(xxxiii)
Cover the ELISA plate with foil or Parafilm and incubate for at least 45 min at 37 °C.
-
(xxxiv)
Remove the coating solution by flicking the plate and tap residual liquid on paper towels.
-
(xxxv)
Wash each well of the ELISA plate six times with 200 µl of washing buffer (see ‘Reagent setup’) and remove remaining fluids by flicking the plate.
-
(xxxvi)
Add 200 µl of blocking buffer (see ‘Reagent setup’) per well.
-
(xxxvii)
Incubate ELISA plate for at least 45 min at 37 °C.
-
(xxxviii)
In the meantime, prepare a dilution series of the supernatants from Step 135A(xxix) as follows:
-
Fill rows B to H of a noncoated Microtest 96-well plate with 100 µl of 1× PBS (supernatant dilution plate).
-
Dilute standard human myeloma IgG1 kappa to 4 µg/ml in 1× PBS and add 120 µl to well A1 of each supernatant dilution plate.
-
Prepare a 1:20 dilution of the HEK293T supernatants with 1× PBS and add 120 µl of each supernatant dilution to one of the remaining wells from A2 to A12.
-
Transfer 50 µl from row A to B and mix with a multichannel pipette. Then, transfer 50 µl of row B to row C, and mix. Proceed accordingly row by row. This will result in a 1:3 dilution series from row to row.
-
-
(xxxix)
Wash each well of the ELISA plate six times with 200 µl of washing buffer (see ‘Reagent setup’) and flick remaining fluids.
-
(xl)
Transfer 50 µl of each well of the supernatant dilution plate to the same well number of the ELISA plate, starting with the lowest concentration in row H.
Critical step
As samples are transferred from low to high concentrations, there is no need to change tips here.
-
(xli)
Incubate ELISA plates for 45 min at room temperature.
-
(xlii)
Dilute HRP-conjugated secondary antibody 1:1,000 in blocking buffer.
-
(xliii)
Wash each well of the ELISA plate six times with 200 µl of washing buffer (see ‘Reagent setup’) and flick remaining fluids.
-
(xliv)
Use a multichannel pipette to add 50 µl of the HRP-conjugated antibody dilution to each well, starting at row H.
Critical step
Do not change pipette tips in between the rows as samples are transferred from low to high concentrations
-
(xlv)
Incubate at room temperature for 45 min.
-
(xlvi)
In the meantime, equilibrate ABTS solution to room temperature.
-
(xlvii)
Wash each well of the ELISA plate six times with 200 µl of washing buffer (see ‘Reagent setup’) and flick remaining fluids.
-
(xlviii)
Start the reaction by adding 100 µl of ABTS solution per well, starting in row H of the ELISA plate.
-
(xlix)
Measure the absorbance at 415 nm wavelength ~4 min after reaction start and determine antibody concentration in cell culture supernatants by comparison with the standard in column A.
-
(l)
Cell culture supernatants containing antibodies can be used for selected downstream applications without further purification (e.g., ELISA or microneutralization assays).
-
(i)
-
(B)
Large-scale antibody production
Midi DNA preparation (low-throughput)
Timing 24 h (days 5–6)
-
(i)
Align sequences of colony PCR products to sequences from second PCR products (Step 68).
-
(ii)
Inoculate liquid cultures for midi-scale plasmid-DNA preparation for each correct pair of heavy and light chains directly from the 96-well liquid culture or respective glycerol stock plate cPCR I/II or III from Step 129 in 50 ml of LB medium.
-
(iii)
Proceed with midi-scale plasmid-DNA preparation according to the manufacturer’s protocol.
-
(iv)
Sequence midi-scale plasmid-DNA with 5′ Ab-sense primer.
Pause point
Midi-scale plasmid-DNA preparations can be stored at −20 °C for up to 3 years.
Transfection of HEK293-6E cells (low-throughput)
Timing 3 h (days 7–14)
-
(v)
Work under a sterile hood with laminar flow.
-
(vi)
Count HEK293-6E suspension cells to be transfected. Cells should be in the range of 8.0 × 105 to 1.2 × 106 per ml for transfection.
-
(vii)
Prepare transfection mixes for different culture volumes in a 50 ml conical tube as follows:
-
Mix heavy- and light-chain plasmids
-
Add DPBS and briefly vortex the suspension
-
Add PEI, vortex for at least 15 s and incubate for 10 min at room temperature
Critical step
For transfection complex formation, mixing and incubation for the indicated time is important.
Composition of HEK293-6E transfection mix
Culture volume (ml)
50
100
240
400
DPBS (ml)
2.25
4.5
10.8
18
Heavy- and light-chain plasmid (µg each)
25
50
120
200
PEI (0.45 mg/ml) (ml)
0.17
0.34
0.82
1.36
-
-
(viii)
Retrieve HEK293-6E suspension cell culture from incubation shaker.
-
(ix)
Add transfection mix under constant agitation to the cell culture flasks.
-
(x)
Incubate the cells for 7 d at 110 rpm, 37 °C and 6.0% CO2 in cell culture incubation shaker.
Protein G–based antibody purification (low-throughput)
Timing 7 h (day 14)
-
(xi)
Transfer cell supernatants from transfected cell cultures from Step 135B(x) into a 50 ml conical tube.
Critical step
Antibody expression level can be estimated by SDS-PAGE prior to purification.
-
(xii)
Centrifuge cell suspension at 3,300g and 4 °C for 10 min.
-
(xiii)
Calculate the amount of protein G beads needed for purification. For transfected HEK293-6E cultures, the expected IgG concentration is <5 mg/50 ml. Binding capacity of protein G sepharose is 1.7 mg of human IgG/100 µl. Add at least 200 µl of beads per 50 ml cell supernatant.
-
(xiv)
Transfer the appropriate amount of protein G sepharose beads into a 50 ml conical plastic tube.
-
(xv)
Add 5 ml of 1× PBS per 200 µl of beads (first wash).
Critical step
Protein G sepharose beads contain 20% of ethanol as a preservative. Wash beads before use with 1× PBS to remove ethanol.
-
(xvi)
Centrifuge at 600g and 4 °C for 5 min (brake turned off).
-
(xvii)
Carefully remove the supernatant by pipetting and repeat the washing step (second wash)
-
(xviii)
Dilute protein G sepharose beads with 1 ml 1× PBS per 200 µl of beads.
-
(xix)
Add ~1 ml of protein G suspension with a serological pipette to the cell supernatants and incubate 2 h at room temperature under constant rotation.
Pause point
Cell supernatants can be incubated with protein G beads for up to 5 d at 4 °C.
-
(xx)
After incubation, centrifuge supernatants at 600g and 4 °C for 15 min (brake turned off).
-
(xxi)
Wash chromatography columns for collection of protein G beads with 5 ml of 70% ethanol.
-
(xxii)
Equilibrate columns with 5 ml of sterile 1× PBS and let them drain by gravity.
-
(xxiii)
Carefully discard supernatant from Step 135B(xx) with a serological pipette.
-
(xxiv)
Aspirate protein G beads with a 5 ml serological pipette and transfer to chromatography columns.
Critical step
Do not let columns run dry.
-
(xxv)
Wash each column 3× with 10 ml of sterile 1× PBS.
-
(xxvi)
Prepare 15 ml conical plastic tubes with 500 µl of 1 M Tris buffer, pH 8 (see ‘Reagent setup’).
-
(xxvii)
Elute columns with 4.5 ml of 0.1 M Glycine buffer pH 3 (see ‘Reagent setup’) directly into plastic tubes containing the 500 µl of 1 M Tris buffer pH 8.
-
(xxviii)
Measure antibody concentrations by UV/Vis spectroscopy against a blank with a 1:10 dilution of Tris in glycine buffer.
-
(xxix)
Only apply eluate with antibody concentrations >20 µg/ml to Amicon Ultra-4 30 K spin filters.
-
(xxx)
To dilute the high concentrations of Tris and glycine buffer, which might interfere with downstream assays for functional characterization of antibodies, fill up each spin column to 15 ml with 1× PBS (first wash).
-
(xxxi)
Spin columns at 3750g until 500–1,000 µl of wash solution remains in the filter.
Critical step
Be careful not to exceed the solubility product of immunoglobulins to prevent precipitation.
-
(xxxii)
Discard flow through and fill up the spin column to 15 ml with 1× PBS (second wash).
-
(xxxiii)
Repeat the wash step until the Tris and glycine content is diluted by >1:200.
-
(xxxiv)
Sterile-filter antibodies with ultrafree-CL columns.
-
(xxxv)
Measure antibody concentrations by UV/Vis spectroscopy.
Pause point
Purified antibody can be stored at 4 °C for up to 6 months.
-
(i)
-
(A)
Troubleshooting
Troubleshooting advice can be found in Table 1.
Timing
-
Stage I: antigen-specific B-cell isolation
-
PBMC collection, Steps 1–3: 2.5 h (day 1)
-
Isolation of CD19+ B cells, Steps 4–17: 1.5 h (day 1)
-
Staining of antigen-specific B cells and single-cell sorting, Steps 18–30: 3 h (day 1)
-
Stage II: high-efficiency BCR amplification
-
Reverse transcription, Steps 31–47: 2 h (day 1)
-
First single-cell PCR for amplification of heavy- and light-chain genes, Steps 48–56: 3 h (day 1)
-
Second single-cell PCR for amplification heavy- and light-chain genes, Steps 57–69: 3 h (day 2)
-
Sequence annotation, analysis and selection for downstream antibody production, Steps 70–72: 2–4 h (day 3)
-
Stage III: high-efficiency BCR cloning
-
Cloning PCR of heavy- and light-chain genes, Steps 73–83: 2 h (day 3)
-
Linearization of expression vector, Steps 84–94: 8–16 h (days 2–3)
-
SLIC of antibody variable regions, Steps 95–117: 4 h (day 3)
-
Colony PCR and sequencing, Steps 118–134: 24 h (day 4)
-
Stage IV: high-throughput and/or large-scale antibody production
-
A
High-throughput antibody production
-
96-well mini DNA preparation (high-throughput), Step 135A(i–xiv): 48 h (days 5–7)
-
Transfection of HEK293T cells (high-throughput), Step 135A(xv–xxx): 3 h for procedure, 96 h incubation time (days 8–12)
-
Human IgG capture ELISA (high-throughput), Step 135A(xxxi–l): 6 h (day 12)
-
-
B
Large-scale antibody production
-
Midi DNA preparations (low-throughput), Step 135B(i–iv): 24 h (days 5 and 6)
-
Transfection of HEK293-6E cells (low-throughput), Step 135B(v–x): 3 h for procedure, 7 d incubation time (days 7–14)
-
Protein G–based antibody purification (low-throughput), Step 135B(xi–xxxv): 7 h for procedure, 24 h incubation time (day 14)
-
-
A
Anticipated results
For the example results presented in this section and in Fig. 3, human blood samples were obtained under a study protocol approved by the IRB of the University of Cologne and respective local IRBs (study protocol 16-054). All study participants provided written informed consent.
For stage (I), we expect a frequency of live single cells in the range of 75–80%. More than 95% of live single cells are CD20+, and 10–20% of these cells are IgG+. The frequency of antigen-specific cells varies between 0.08% and 1.4%. For stages (II) and (III), exemplary agarose gel electrophoresis analyses for second, cloning and colony PCR are depicted. Green, white and black squares represent amplified, unamplified and control or empty wells, respectively. HC, KC and LC represent heavy, kappa and lambda chain, respectively. For second PCR, we expect a coverage of ≥90% for the amplification of heavy and >85% for light chains. Typically, ≥85% of paired sequences are amplified and 5–10% of heavy chains are amplified with an IgK and IgL light chain. For cloning PCR, we anticipate coverages of ≥90% for amplification of heavy, ≥85% for kappa and ≥80% for lambda light chains. Typically, ≥80% of paired sequences are amplified. For colony PCR, we expect coverages of ≥95% for heavy, >90% for kappa and ≥75% for lambda light chains, when three bacterial clones are picked for analysis. We expect a coverage of ≥80% for the amplification of paired sequences. For stage (IV), a representative human IgG capture ELISA of supernatant dilutions is shown for the high-throughput approach. Dot plots depict yields and concentrations of single wells (A, high-throughput) or shaking flasks (B, large-scale) with median and interquartile ranges. High-throughput production in 48-well format yields up to 20 µg (mean: 4.2 µg, median: 2.6 µg) and concentrations up to 50 µg/ml (mean: 10.5 µg/ml, median: 6.5 µg/ml). Large-scale production yields up to 2 mg (mean: 0.88 mg, median: 0.89 mg) and concentrations up to 20 mg/ml (mean: 6.25 mg/ml, median: 5.68 mg/ml).
Isolation of antigen-specific B cells
From 1 ml of whole blood, ~1 × 106 PBMCs are isolated. Using CD19 microbeads, average purification efficiencies of ~75% are achieved with an average cell viability of 95%. Among isolated PBMCs, ~8% of cells account for CD19+ B cells, of which 11% are IgG+. Frequencies for bait-specific B cells strongly depend on the bait protein and can vary from ~0.3% (Ebola virus or SARS-CoV-2 specific sorts) to 0.08% (HIV-1 BG505 SOSIP.664 gp140 specific sorts) (Fig. 3, stage I of the procedure). All values have been determined by cell counting and FACS analysis.
Optimized BCR amplification
Representative agarose gel electrophoresis analyses for PCR reactions are depicted in Fig. 3 (stages II and III of the procedure). For the second single-cell PCR, we expect an efficiency of ≥90% for the amplification of heavy and ≥85% for the amplification of light chains. Typically, approximately ≥85% of corresponding heavy- and light-chain pairs are amplified. In some cases, ~5–10% of heavy chains are amplified with an IgK and IgL light chain. In up to 20% of these cases, both light-chain alleles are functional.
Optimized BCR cloning
Representative agarose gel electrophoresis analyses for PCR reactions are depicted in Fig. 3 (stages II and III of the procedure). For the cloning PCR, we expect an efficiency of ≥90% for the amplification of heavy, ≥85% for kappa and ≥80% for lambda light chains. Typically, we expect a coverage of ≥80% for the amplification of corresponding heavy- and light-chain pairs. Lambda light-chain amplification can be repeated in separate first single-cell PCR reactions prior to cloning PCR, if low coverage occurs (described in Troubleshooting). After SLIC, transformed bacterial clones are analyzed for correct insertion of the respective V region into expression vectors by colony PCR. Colony PCR efficiencies vary from 80% to 90% for the amplification of heavy, 80% to 90% for kappa and 60% to 65% for lambda light chains when only a single bacterial colony is analyzed. When three bacterial clones for each cloning reaction are analyzed, ≥95% of heavy, ≥90% of kappa and ≥75% of lambda light chains are amplified in the colony PCR. We expect a coverage of ≥80% for the amplification of corresponding heavy- and light-chain pairs.
High-throughput and large-scale antibody production
According to the respective experimental objectives, antibodies can either be produced according to a high- or low-throughput approach. High- and low-throughput approaches differ considerably in yields and concentrations of produced antibodies (Fig. 3, stage IV of the procedure).
Transfection of HEK293T cells (high-throughput)
The mean concentration and yield of produced antibodies are approximately 32 µg/ml (median = 31 µg/ml, 25% percentile = 26 µg/ml, 75% percentile = 38 µg/ml) and 6.3 µg (median = 6.2 µg, 25% percentile = 5.1 µg, 75% percentile = 7.7 µg) for transfections in a 96-well format or 10.5 µg/ml (median = 6.5 µg/ml, 25% percentile = 2.1 µg/ml, 75% percentile = 16.4 µg/ml) and 4.2 µg (median = 2.6 µg, 25% percentile = 0.8 µg, 75% percentile = 6.6 µg) in a 48-well format, respectively. Around 89% of transfections produce antibodies with a concentration ≥1 µg/ml in 400 µl of supernatant in a 48-well format.
Transfection of HEK293-6E cells (low-throughput)
For the low-throughput approach, the protocol was optimized for transfection of HEK293-6E suspension cells in a 50 ml culture. Concentration and yields are typically in the range of 2–20 mg/ml and 0.4–2.0 mg, respectively.
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
Source data are provided with this paper.
References
Grilo, A. L. & Mantalaris, A. The increasingly human and profitable monoclonal antibody market. Trends Biotechnol 37, 9–16 (2019).
Kaplon, H., Muralidharan, M., Schneider, Z. & Reichert, J. M. Antibodies to watch in 2020. MAbs 12, 1703531 (2020).
Corti, D. et al. Cross-neutralization of four paramyxoviruses by a human monoclonal antibody. Nature 501, 439–443 (2013).
Smith, S. A. et al. Isolation and characterization of broad and ultrapotent human monoclonal antibodies with therapeutic activity against Chikungunya virus. Cell Host Microbe 18, 86–95 (2015).
Stettler, K. et al. Specificity, cross-reactivity, and function of antibodies elicited by Zika virus infection. Science 353, 823–826 (2016).
Mendoza, P. et al. Combination therapy with anti-HIV-1 antibodies maintains viral suppression. Nature 561, 479–484 (2018).
Bournazos, S. et al. Broadly neutralizing anti-HIV-1 antibodies require Fc effector functions for in vivo activity. Cell 158, 1243–1253 (2014).
van Erp, E. A., Luytjes, W., Ferwerda, G. & van Kasteren, P. B. Fc-mediated antibody effector functions during respiratory syncytial virus infection and disease. Front. Immunol. 10, 548 (2019).
Asokan, M. et al. Fc-mediated effector function contributes to the in vivo antiviral effect of an HIV neutralizing antibody. Proc. Natl Acad. Sci. USA 117, 18754–18763 (2020).
Keizer, R. J., Huitema, A. D. R., Schellens, J. H. M. & Beijnen, J. H. Clinical pharmacokinetics of therapeutic monoclonal antibodies. Clin. Pharmacokinet. 49, 493–507 (2010).
Ryman, J. T. & Meibohm, B. Pharmacokinetics of monoclonal antibodies. CPT Pharmacometrics Syst. Pharmacol. 6, 576–588 (2017).
Cohen, Y. Z. et al. Safety, pharmacokinetics, and immunogenicity of the combination of the broadly neutralizing anti-HIV-1 antibodies 3BNC117 and 10-1074 in healthy adults: A randomized, phase 1 study. PLoS ONE 14, e0219142 (2019).
Pelegrin, M., Naranjo-Gomez, M. & Piechaczyk, M. Antiviral monoclonal antibodies: can they be more than simple neutralizing agents? Trends Microbiol. 23, 653–665 (2015).
Panda, S. & Ding, J. L. Natural antibodies bridge innate and adaptive immunity. J. Immunol. 194, 13–20 (2015).
Zuniga, E. I., Macal, M., Lewis, G. M. & Harker, J. A. Innate and adaptive immune regulation during chronic viral infections. Annu. Rev. Virol. 2, 573–597 (2015).
Küppers, R., Zhao, M., Hansmann, M. L. & Rajewsky, K. Tracing B cell development in human germinal centres by molecular analysis of single cells picked from histological sections. EMBO J. 12, 4955–4967 (1993).
Wardemann, H. et al. Predominant autoantibody production by early human B cell precursors. Science 301, 1374–1377 (2003).
Smith, K. et al. Rapid generation of fully human monoclonal antibodies specific to a vaccinating antigen. Nat. Protoc. 4, 372–384 (2009).
von Boehmer, L. et al. Sequencing and cloning of antigen-specific antibodies from mouse memory B cells. Nat. Protoc. 11, 1908–1923 (2016).
Mascola, J. R. et al. Protection of macaques against pathogenic simian/human immunodeficiency virus 89.6PD by passive transfer of neutralizing antibodies. J. Virol. 73, 4009–4018 (1999).
Mascola, J. R. et al. Protection of macaques against vaginal transmission of a pathogenic HIV-1/SIV chimeric virus by passive infusion of neutralizing antibodies. Nat. Med. 6, 207–210 (2000).
Balazs, A. B. et al. Antibody-based protection against HIV infection by vectored immunoprophylaxis. Nature 481, 81–84 (2011).
Schepens, B. et al. Nanobodies® specific for respiratory syncytial virus fusion protein protect against infection by inhibition of fusion. J. Infect. Dis. 204, 1692–1701 (2011).
Detalle, L. et al. Generation and characterization of ALX-0171, a potent novel therapeutic nanobody for the treatment of respiratory syncytial virus infection. Antimicrob. Agents Chemother. 60, 6–13 (2016).
Mire, C. E. et al. Human-monoclonal-antibody therapy protects nonhuman primates against advanced Lassa fever. Nat. Med. 23, 1146–1149 (2017).
Paules, C. I. et al. The hemagglutinin A stem antibody MEDI8852 prevents and controls disease and limits transmission of pandemic influenza viruses. J. Infect. Dis. 216, 356–365 (2017).
Van Heeke, G. et al. Nanobodies® as inhaled biotherapeutics for lung diseases. Pharmacol. Ther. 169, 47–56 (2017).
Walker, L. M. & Burton, D. R. Passive immunotherapy of viral infections: ‘super-antibodies’ enter the fray. Nat. Rev. Immunol. 18, 297–308 (2018).
Graham, B. S., Gilman, M. S. A. & McLellan, J. S. Structure-based vaccine antigen design. Annu. Rev. Med. 70, 91–104 (2019).
Kreer, C., Gruell, H., Mora, T., Walczak, A. M. & Klein, F. Exploiting B cell receptor analyses to inform on HIV-1 vaccination strategies. Vaccines 8, 13 (2020).
Gaebler, C. et al. Isolation of HIV-1-reactive antibodies using cell surface-expressed gp160Δc(BaL.). J. Immunol. Methods 397, 47–54 (2013).
Corti, D. et al. Prophylactic and postexposure efficacy of a potent human monoclonal antibody against MERS coronavirus. Proc. Natl Acad. Sci. USA 112, 10473–10478 (2015).
Xiao, X. et al. A novel antibody discovery platform identifies anti-influenza A broadly neutralizing antibodies from human memory B cells. MAbs 8, 916–927 (2016).
Ehrhardt, S. A. et al. Polyclonal and convergent antibody response to Ebola virus vaccine rVSV-ZEBOV. Nat. Med. 25, 1589–1600 (2019).
Schoofs, T. et al. Broad and potent neutralizing antibodies recognize the silent face of the HIV envelope. Immunity 50, 1513–1529.e9 (2019).
Schommers, P. et al. Restriction of HIV-1 escape by a highly broad and potent neutralizing antibody. Cell 180, 471–489.e22 (2020).
Kreer, C. et al. Longitudinal isolation of potent near-germline SARS-CoV-2-neutralizing antibodies from COVID-19 patients. Cell 182, 843–854.e12 (2020).
Rogers, T. F. et al. Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science 369, 956–963 (2020).
Babcook, J. S., Leslie, K. B., Olsen, O. A., Salmon, R. A. & Schrader, J. W. A novel strategy for generating monoclonal antibodies from single, isolated lymphocytes producing antibodies of defined specificities. Proc. Natl Acad. Sci. USA 93, 7843–7848 (1996).
Harding, F. A., Stickler, M. M., Razo, J. & DuBridge, R. B. The immunogenicity of humanized and fully human antibodies: residual immunogenicity resides in the CDR regions. MAbs 2, 256–265 (2010).
Laffleur, B., Pascal, V., Sirac, C. & Cogné, M. Production of human or humanized antibodies in mice. Methods Mol. Biol. 901, 149–159 (2012).
Tiller, T. et al. Efficient generation of monoclonal antibodies from single human B cells by single cell RT-PCR and expression vector cloning. J. Immunol. Methods 329, 112–124 (2008).
Joyce, M. G. et al. Vaccine-induced antibodies that neutralize group 1 and group 2 influenza A viruses. Cell 166, 609–623 (2016).
Robbiani, D. F. et al. Recurrent potent human neutralizing antibodies to Zika virus in Brazil and Mexico. Cell 169, 597–609.e11 (2017).
Theobald, S. J. et al. Repertoire characterization and validation of gB-specific human IgGs directly cloned from humanized mice vaccinated with dendritic cells and protected against HCMV. PLoS Pathog. 16, e1008560 (2020).
Döring, M., Kreer, C., Lehnen, N., Klein, F. & Pfeifer, N. Modeling the amplification of immunoglobulins through machine learning on sequence-specific features. Sci. Rep. 9, 10748 (2019).
Kreer, C. et al. openPrimeR for multiplex amplification of highly diverse templates. J. Immunol. Methods 480, 112752 (2020).
Scheid, J. F. et al. Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science 333, 1633–1637 (2011).
Liao, H.-X. et al. High-throughput isolation of immunoglobulin genes from single human B cells and expression as monoclonal antibodies. J. Virol. Methods 158, 171–179 (2009).
Sanders, R. W. et al. A next-generation cleaved, soluble HIV-1 Env trimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLoS Pathog. 9, e1003618 (2013).
Lan, J. et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 581, 215–220 (2020).
Pallesen, J. et al. Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen. Proc. Natl Acad. Sci. USA 114, E7348–E7357 (2017).
Scherer, E. M. et al. Characteristics of memory B cells elicited by a highly efficacious HPV vaccine in subjects with no pre-existing immunity. PLoS Pathog. 10, e1004461 (2014).
Bornholdt, Z. A. et al. Isolation of potent neutralizing antibodies from a survivor of the 2014 Ebola virus outbreak. Science 351, 1078–1083 (2016).
Gilman, M. S. A. et al. Rapid profiling of RSV antibody repertoires from the memory B cells of naturally infected adult donors. Sci. Immunol. 1, eaaj1879 (2016).
Andrews, S. F. et al. Preferential induction of cross-group influenza A hemagglutinin stem-specific memory B cells after H7N9 immunization in humans. Sci. Immunol. 2, eaan2676 (2017).
Dussupt, V. et al. Potent Zika and dengue cross-neutralizing antibodies induced by Zika vaccination in a dengue-experienced donor. Nat. Med. 26, 228–235 (2020).
Ma, K. et al. Multiple functions of B cells in the pathogenesis of systemic lupus erythematosus. Int. J. Mol. Sci. 20, 6021 (2019).
Tipton, C. M., Hom, J. R., Fucile, C. F., Rosenberg, A. F. & Sanz, I. Understanding B-cell activation and autoantibody repertoire selection in systemic lupus erythematosus: a B-cell immunomics approach. Immunol. Rev. 284, 120–131 (2018).
Marston, B., Palanichamy, A. & Anolik, J. H. B cells in the pathogenesis and treatment of rheumatoid arthritis. Curr. Opin. Rheumatol. 22, 307–315 (2010).
Sharonov, G. V., Serebrovskaya, E. O., Yuzhakova, D. V., Britanova, O. V. & Chudakov, D. M. B cells, plasma cells and antibody repertoires in the tumour microenvironment. Nat. Rev. Immunol. 20, 294–307 (2020).
Scheid, J. F. et al. A method for identification of HIV gp140 binding memory B cells in human blood. J. Immunol. Methods 343, 65–67 (2009).
Huse, W. D. et al. Generation of a large combinatorial library of the immunoglobulin repertoire in phage lambda. Science 246, 1275–1281 (1989).
Boder, E. T. & Wittrup, K. D. Yeast surface display for screening combinatorial polypeptide libraries. Nat. Biotechnol. 15, 553–557 (1997).
Wiesner, M. et al. Conditional immortalization of human B cells by CD40 ligation. PLoS ONE 3, e1464 (2008).
Huang, J. et al. Identification of a CD4-binding-site antibody to HIV that evolved near-pan neutralization breadth. Immunity 45, 1108–1121 (2016).
Wec, A. Z. et al. Antibodies from a human survivor define sites of vulnerability for broad protection against ebolaviruses. Cell 169, 878–890.e15 (2017).
McCafferty, J., Griffiths, A. D., Winter, G. & Chiswell, D. J. Phage antibodies: filamentous phage displaying antibody variable domains. Nature 348, 552–554 (1990).
Kwakkenbos, M. J., van Helden, P. M., Beaumont, T. & Spits, H. Stable long-term cultures of self-renewing B cells and their applications. Immunol. Rev. 270, 65–77 (2016).
Doria-Rose, N. A. et al. New member of the V1V2-directed CAP256-VRC26 lineage that shows increased breadth and exceptional potency. J. Virol. 90, 76–91 (2016).
Kwakkenbos, M. J. et al. Generation of stable monoclonal antibody-producing B cell receptor-positive human memory B cells by genetic programming. Nat. Med. 16, 123–128 (2010).
Corti, D. et al. A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science 333, 850–856 (2011).
Kwong, P. D. & Mascola, J. R. Human antibodies that neutralize HIV-1: identification, structures, and B cell ontogenies. Immunity 37, 412–425 (2012).
Sok, D. et al. Rapid elicitation of broadly neutralizing antibodies to HIV by immunization in cows. Nature 548, 108–111 (2017).
Zhao, X. et al. Immunization-elicited broadly protective antibody reveals Ebolavirus fusion loop as a site of vulnerability. Cell 169, 891–904.e15 (2017).
Ye, J., Ma, N., Madden, T. L. & Ostell, J. M. IgBLAST: an immunoglobulin variable domain sequence analysis tool. Nucleic Acids Res. 41, W34–W40 (2013).
Lefranc, M. P. et al. IMGT, the international ImMunoGeneTics database. Nucleic Acids Res. 27, 209–212 (1999).
Hsieh, C.-L. et al. Structure-based design of prefusion-stabilized SARS-CoV-2 spikes. Science 369, 1501–1505 (2020).
Giudicelli, V., Chaume, D. & Lefranc, M.-P. IMGT/V-QUEST, an integrated software program for immunoglobulin and T cell receptor V-J and V-D-J rearrangement analysis. Nucleic Acids Res. 32, W435–W440 (2004).
Acknowledgements
We thank M. Nussenzweig and members of the Nussenzweig lab for valuable support as well as intital protocols on single B-cell cloning and antibody production. We thank all members of the Klein lab for helpful discussion. This work was funded by grants from the European Research Council (ERC-StG639961, to F.K.), the German Centre for Infection Research (DZIF, to F.K., H.G. and L.G.), the Bill and Melinda Gates Foundation (BMGF, INV-002143 to F.K.), the German Research Foundation (DFG) (CRC 1279 and CRC 1310 to F.K.) and the COVIM: NaFoUniMedCovid19 (FKZ: 01KX2021 to F.K.).
Author information
Authors and Affiliations
Contributions
L.G., C.K. and F.K. conceptualized the workflow. L.G, C.K, M.S.E., M.Z., N.L., H.G., P.S. and J.P. established protocols and performed experiments. F.K. and C.K. supervised the development of the protocol. L.G., C.K. and F.K. wrote the manuscript. All authors were involved in editing of the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Protocols thanks the anonymous reviewers for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key references using this protocol
Ehrhardt, S. A. et al. Nat. Med. 25, 1589–1600 (2019): https://doi.org/10.1038/s41591-019-0602-4
Schommers, P. et al. Cell 180, 471–489.e22 (2020): https://doi.org/10.1016/j.cell.2020.01.010
Kreer, C. et al. Cell 182, 843–854.e12 (2020): https://doi.org/10.1016/j.cell.2020.06.044
Supplementary information
Supplementary Information
Supplementary Tables 1–9.
Source data
Source Data Fig. 3
Unprocessed gel pictures.
Source Data Fig. 3
Statistical source data.
Rights and permissions
About this article
Cite this article
Gieselmann, L., Kreer, C., Ercanoglu, M.S. et al. Effective high-throughput isolation of fully human antibodies targeting infectious pathogens. Nat Protoc 16, 3639–3671 (2021). https://doi.org/10.1038/s41596-021-00554-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-021-00554-w
This article is cited by
-
Broadly neutralizing human antibodies against Omicron subvariants of SARS-CoV-2
Journal of Biomedical Science (2023)
-
A rapid cell-free expression and screening platform for antibody discovery
Nature Communications (2023)
-
Dynamics and durability of HIV-1 neutralization are determined by viral replication
Nature Medicine (2023)
-
Neutralizing and enhancing antibodies against SARS-CoV-2
Inflammation and Regeneration (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.