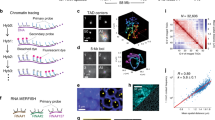
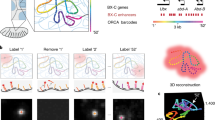
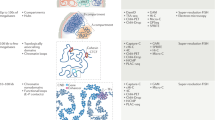
Abstract
The genome is hierarchically organized into several 3D architectures, including chromatin loops, domains, compartments and regions associated with nuclear lamina and nucleoli. Changes in these architectures have been associated with normal development, aging and a wide range of diseases. Despite its critical importance, understanding how the genome is spatially organized in single cells, how organization varies in different cell types in mammalian tissue and how organization affects gene expression remains a major challenge. Previous approaches have been limited by a lack of capacity to directly trace chromatin folding in 3D and to simultaneously measure genomic organization in relation to other nuclear components and gene expression in the same single cells. We have developed an image-based 3D genomics technique termed ‘chromatin tracing’, which enables direct 3D tracing of chromatin folding along individual chromosomes in single cells. More recently, we also developed multiplexed imaging of nucleome architectures (MINA), which enables simultaneous measurements of multiscale chromatin folding, associations of genomic regions with nuclear lamina and nucleoli and copy numbers of numerous RNA species in the same single cells in mammalian tissue. Here, we provide detailed protocols for chromatin tracing in cell lines and MINA in mammalian tissue, which take 3–4 d for experimental work and 2–3 d for data analysis. We expect these developments to be broadly applicable and to affect many lines of research on 3D genomics by depicting multiscale genomic architectures associated with gene expression, in different types of cells and tissue undergoing different biological processes.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
An example raw dataset of two imaging fields is downloadable from https://campuspress.yale.edu/wanglab/mina-analyst/. The full datasets used to generate Figs. 7 and 8 are not uploaded online because of the prohibitively large size. The full datasets are available from the corresponding author upon request. Source data are provided with this paper.
Code availability
The ProbeDealer package can be downloaded from https://campuspress.yale.edu/wanglab/probedealer. The MinaAnalyst package can be downloaded from https://campuspress.yale.edu/wanglab/mina-analyst/.
References
Finn, E. H. & Misteli, T. Molecular basis and biological function of variability in spatial genome organization. Science 365, eaaw9498 (2019).
Gorkin, D. U., Leung, D. & Ren, B. The 3D genome in transcriptional regulation and pluripotency. Cell Stem Cell 14, 762–775 (2014).
Levine, M., Cattoglio, C. & Tjian, R. Looping back to leap forward: transcription enters a new era. Cell 157, 13–25 (2014).
Dekker, J. & Mirny, L. The 3D genome as moderator of chromosomal communication. Cell 164, 1110–1121 (2016).
Bickmore, W. A. & van Steensel, B. Genome architecture: domain organization of interphase chromosomes. Cell 152, 1270–1284 (2013).
Cremer, T. & Cremer, M. Chromosome territories. Cold Spring Harb. Perspect. Biol. 2, a003889 (2010).
Bonora, G., Plath, K. & Denholtz, M. A mechanistic link between gene regulation and genome architecture in mammalian development. Curr. Opin. Genet. Dev. 27, 92–101 (2014).
Pope, B. D. et al. Topologically associating domains are stable units of replication-timing regulation. Nature 515, 402–405 (2014).
van Steensel, B. & Furlong, E. E. M. The role of transcription in shaping the spatial organization of the genome. Nat. Rev. Mol. Cell Biol. 20, 327–337 (2019).
Zhang, Y. et al. Spatial organization of the mouse genome and its role in recurrent chromosomal translocations. Cell 148, 908–921 (2012).
Senigl, F. et al. Topologically associated domains delineate susceptibility to somatic hypermutation. Cell Rep 29, 3902–3915.e8 (2019).
Misteli, T. & Soutoglou, E. The emerging role of nuclear architecture in DNA repair and genome maintenance. Nat. Rev. Mol. Cell Biol. 10, 243–254 (2009).
Akdemir, K. C. et al. Disruption of chromatin folding domains by somatic genomic rearrangements in human cancer. Nat. Genet. 52, 294–305 (2020).
Lieberman-Aiden, E. et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326, 289–293 (2009).
Rao, S. S. P. et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159, 1665–1680 (2014).
Wang, Q., Sun, Q., Czajkowsky, D. M. & Shao, Z. Sub-kb Hi-C in D. melanogaster reveals conserved characteristics of TADs between insect and mammalian cells. Nat. Commun. 9, 188 (2018).
Wang, S. et al. Spatial organization of chromatin domains and compartments in single chromosomes. Science 353, 598–602 (2016).
Liu, M. et al. Multiplexed imaging of nucleome architectures in single cells of mammalian tissue. Nat. Commun. 11, 2907 (2020).
Bintu, B. et al. Super-resolution chromatin tracing reveals domains and cooperative interactions in single cells. Science 362, eaau1783 (2018).
Sawh, A. N. et al. Lamina-dependent stretching and unconventional chromosome compartments in early C. elegans embryos. Mol. Cell 78, 96–111.e6 (2020).
Nir, G. et al. Walking along chromosomes with super-resolution imaging, contact maps, and integrative modeling. PLoS Genet. 14, e1007872 (2018).
Mateo, L. J. et al. Visualizing DNA folding and RNA in embryos at single-cell resolution. Nature 568, 49–54 (2019).
Cardozo Gizzi, A. M. et al. Microscopy-based chromosome conformation capture enables simultaneous visualization of genome organization and transcription in intact organisms. Mol. Cell 74, 212–222.e5 (2019).
Su, J.-H., Zheng, P., Kinrot, S. S., Bintu, B. & Zhuang, X. Genome-scale imaging of the 3D organization and transcriptional activity of chromatin. Cell 182, 1641–1659.e26 (2020).
Hu, M. & Wang, S. Chromatin tracing: imaging 3D genome and nucleome. Trends Cell Biol. 31, 5–8 (2021).
Plath, K., Mlynarczyk-Evans, S., Nusinow, D. A. & Panning, B. Xist RNA and the mechanism of X chromosome inactivation. Annu. Rev. Genet. 36, 233–278 (2002).
Galupa, R. & Heard, E. X-chromosome inactivation: new insights into cis and trans regulation. Curr. Opin. Genet. Dev. 31, 57–66 (2015).
Lessing, D., Anguera, M. C. & Lee, J. T. X chromosome inactivation and epigenetic responses to cellular reprogramming. Annu. Rev. Genomics Hum. Genet. 14, 85–110 (2013).
Guelen, L. et al. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature 453, 948–951 (2008).
Németh, A. et al. Initial genomics of the human nucleolus. PLOS Genet 6, e1000889 (2010).
van Koningsbruggen, S. et al. High-resolution whole-genome sequencing reveals that specific chromatin domains from most human chromosomes associate with nucleoli. Mol. Biol. Cell 21, 3735–3748 (2010).
Chen, K. H., Boettiger, A. N., Moffitt, J. R., Wang, S. & Zhuang, X. Spatially resolved, highly multiplexed RNA profiling in single cells. Science 348, aaa6090 (2015).
Moffitt, J. R. et al. Molecular, spatial, and functional single-cell profiling of the hypothalamic preoptic region. Science 362, eaau5324 (2018).
Xia, C., Fan, J., Emanuel, G., Hao, J. & Zhuang, X. Spatial transcriptome profiling by MERFISH reveals subcellular RNA compartmentalization and cell cycle-dependent gene expression. Proc. Natl Acad. Sci. USA 116, 19490–19499 (2019).
Boyle, S., Rodesch, M. J., Halvensleben, H. A., Jeddeloh, J. A. & Bickmore, W. A. Fluorescence in situ hybridization with high-complexity repeat-free oligonucleotide probes generated by massively parallel synthesis. Chromosome Res. 19, 901–909 (2011).
Yamada, N. A. et al. Visualization of fine-scale genomic structure by oligonucleotide-based high-resolution FISH. Cytogenet. Genome Res. 132, 248–254 (2011).
Beliveau, B. J. et al. Versatile design and synthesis platform for visualizing genomes with Oligopaint FISH probes. Proc. Natl Acad. Sci. USA 109, 21301–21306 (2012).
Hu, M. et al. ProbeDealer is a convenient tool for designing probes for highly multiplexed fluorescence in situ hybridization. Sci. Rep. 10, 22031 (2020).
Farack, L. & Itzkovitz, S. Protocol for single-molecule fluorescence in situ hybridization for intact pancreatic tissue. STAR Protoc 1, 100007 (2020).
Moffitt, J. R. et al. High-performance multiplexed fluorescence in situ hybridization in culture and tissue with matrix imprinting and clearing. Proc. Natl Acad. Sci. USA 113, 14456–14461 (2016).
Xia, C., Babcock, H. P., Moffitt, J. R. & Zhuang, X. Multiplexed detection of RNA using MERFISH and branched DNA amplification. Sci. Rep. 9, 7721 (2019).
Stevens, T. J. et al. 3D structures of individual mammalian genomes studied by single-cell Hi-C. Nature 544, 59–64 (2017).
Nagano, T. et al. Single-cell Hi-C reveals cell-to-cell variability in chromosome structure. Nature 502, 59–64 (2013).
Tan, L., Xing, D., Chang, C.-H., Li, H. & Xie, X. S. Three-dimensional genome structures of single diploid human cells. Science 361, 924–928 (2018).
Dey, S. S., Kester, L., Spanjaard, B., Bienko, M. & van Oudenaarden, A. Integrated genome and transcriptome sequencing of the same cell. Nat. Biotechnol. 33, 285–289 (2015).
Macaulay, I. C. et al. G&T-seq: parallel sequencing of single-cell genomes and transcriptomes. Nat. Methods 12, 519–522 (2015).
Moffitt, J. R. et al. High-throughput single-cell gene-expression profiling with multiplexed error-robust fluorescence in situ hybridization. Proc. Natl Acad. Sci. USA 113, 11046–11051 (2016).
Rouillard, J.-M. OligoArray 2.0: design of oligonucleotide probes for DNA microarrays using a thermodynamic approach. Nucleic Acids Res. 31, 3057–3062 (2003).
Beliveau, B. J. et al. OligoMiner provides a rapid, flexible environment for the design of genome-scale oligonucleotide in situ hybridization probes. Proc. Natl Acad. Sci. USA 115, E2183–E2192 (2018).
Passaro, M. et al. OligoMinerApp: a web-server application for the design of genome-scale oligonucleotide in situ hybridization probes through the flexible OligoMiner environment. Nucleic Acids Res. 48, W332–W339 (2020).
Moffitt, J. R. & Zhuang, X. RNA imaging with multiplexed error-robust fluorescence in situ hybridization (MERFISH). Methods Enzymol. 572, 1–49 (2016).
Rasnik, I., McKinney, S. A. & Ha, T. Nonblinking and long-lasting single-molecule fluorescence imaging. Nat. Methods 3, 891 (2006).
Acknowledgements
S.W. is supported by NIH Director’s New Innovator Award 1DP2GM137414, NCI grant 1R33CA251037, NHGRI grant 1R01HG011245 and 4DN NCI grant 1U01CA260701. This work is partially supported by NHGRI grant 1R01HG011245. M.H. and Y.Cheng are supported by a China Scholarship Council (CSC) Grant. J.S.D.R. is supported by an NIH Predoctoral Training Grant (2T32GM007499). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Author information
Authors and Affiliations
Contributions
S.W. conceived and developed the chromatin-tracing technique with help from others and conceived the MINA technique. M.L. performed experiments in developing MINA with help from others. M.H., B.Y. and S.W. built the ProbeDealer package with help from others. S.W. built the MinaAnalyst package. M.L. and S.W. wrote the manuscript with inputs from B.Y., M.H., J.S.D.R., Y. Chen, S.J. and Y. Cheng.
Corresponding author
Ethics declarations
Competing interests
S.W. is one of the inventors on a patent applied for by Harvard University related to MERFISH. The other authors declare no competing interests.
Additional information
Peer review information Nature Protocols thanks Vadim Backman and Srinjan Basu for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key references using this protocol
Wang, S. et al. Science 353, 598–602 (2016): https://doi.org/10.1126/science.aaf8084
Liu, M. et al. Nat. Commun. 11, 2907 (2020): https://doi.org/10.1038/s41467-020-16732-5
Hu, M. et al. Sci. Rep. 10, 22031 (2020): https://doi.org/10.1038/s41598-020-76439-x
Key data used in this protocol
Liu, M. et al. Nat. Commun. 11, 2907 (2020): https://doi.org/10.1038/s41467-020-16732-5
Supplementary information
Supplementary Table 1
Sequences of the forward primers, reverse primers and reverse transcription primers for primary probe synthesis
Supplementary Table 2
Sequences of Alexa Fluor 750–labeled MERFISH secondary probes, Alexa Fluor 647–labeled chromatin-tracing secondary probes and ATTO 565–labeled chromatin-tracing secondary probes
Source data
Source Data Fig. 2
Unprocessed gels.
Source Data Fig. 7
Statistical source data.
Source Data Fig. 8a
Unprocessed images.
Source Data Fig. 8b–e
Statistical source data.
Rights and permissions
About this article
Cite this article
Liu, M., Yang, B., Hu, M. et al. Chromatin tracing and multiplexed imaging of nucleome architectures (MINA) and RNAs in single mammalian cells and tissue. Nat Protoc 16, 2667–2697 (2021). https://doi.org/10.1038/s41596-021-00518-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-021-00518-0
This article is cited by
-
pyHiM: a new open-source, multi-platform software package for spatial genomics based on multiplexed DNA-FISH imaging
Genome Biology (2024)
-
Polycomb repression of Hox genes involves spatial feedback but not domain compaction or phase transition
Nature Genetics (2024)
-
The spatial organization of transcriptional control
Nature Reviews Genetics (2023)
-
TAD-like single-cell domain structures exist on both active and inactive X chromosomes and persist under epigenetic perturbations
Genome Biology (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.