Abstract
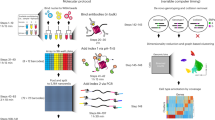
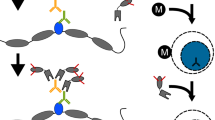
Determining chromatin-associated protein localization across the genome has provided insight into the functions of DNA-binding proteins and their connections to disease. However, established protocols requiring large quantities of cell or tissue samples currently limit applications for clinical and biomedical research in this field. Furthermore, most technologies have been optimized to assess abundant histone protein localization, prohibiting the investigation of nonhistone protein localization in low cell numbers. We recently described a protocol to profile chromatin-associated protein localization in as low as one cell: ultra-low-input cleavage under targets and release using nuclease (uliCUT&RUN). Optimized from chromatin immunocleavage and CUT&RUN, uliCUT&RUN is a tethered enzyme-based protocol that utilizes a combination of recombinant protein, antibody recognition and stringent purification to selectively target proteins of interest and isolate the associated DNA. Performed in native conditions, uliCUT&RUN profiles protein localization to chromatin with low input and high precision. Compared with other profiling technologies, uliCUT&RUN can determine nonhistone protein chromatin occupancies in low cell numbers, permitting the investigation into the molecular functions of a range of DNA-binding proteins within rare samples. From sample preparation to sequencing library submission, the uliCUT&RUN protocol takes <2 d to perform, with the accompanying data analysis timeline dependent on experience level.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Code availability
Code for sequencing data analysis is available at https://github.com/bjp86/Patty_uliCUTandRUNAnalysis.
References
Spitz, F. & Furlong, E. E. M. Transcription factors: from enhancer binding to developmental control. Nat. Rev. Genet. 13, 613–626 (2012).
Villar, D., Flicek, P. & Odom, D. T. Evolution of transcription factor binding in metazoans—mechanisms and functional implications. Nat. Rev. Genet. 15, 221–233 (2014).
Mitsis, T. et al. Transcription factors and evolution: an integral part of gene expression (Review). World Acad. Sci. J. 3–8 (2020)
Andersson, R. & Sandelin, A. Determinants of enhancer and promoter activities of regulatory elements. Nat. Rev. Genet. 21, 71–87 (2020).
Whyte, W. A. et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 153, 307–319 (2013).
Lambert, S. A. et al. The human transcription factors. Cell 172, 650–665 (2018).
Zaret, K. S. & Mango, S. E. Pioneer transcription factors, chromatin dynamics, and cell fate control. Curr. Opin. Genet. Dev. 37, 76–81 (2016).
Iwafuchi-Doi, M. & Zaret, K. S. Cell fate control by pioneer transcription factors. Dev 143, 1833–1837 (2016).
Hu, P., Zhang, W., Xin, H. & Deng, G. Single cell isolation and analysis. Front. Cell Dev. Biol. 4, 1–12 (2016).
Pijuan-Sala, B. et al. A single-cell molecular map of mouse gastrulation and early organogenesis. Nature 566, 490–495 (2019).
Cheng, S. et al. Single-cell RNA-seq reveals cellular heterogeneity of pluripotency transition and X chromosome dynamics during early mouse development. Cell Rep 26, 2593–2607.e3 (2019).
Petropoulos, S. et al. Single-cell RNA-seq reveals lineage and X chromosome dynamics in human preimplantation embryos. Cell 165, 1012–1026 (2016).
Hainer, S. J., Bošković, A., McCannell, K. N., Rando, O. J. & Fazzio, T. G. Profiling of pluripotency factors in single cells and early embryos. Cell 177, 1319–1329.e11 (2019).
Hainer, S. J. & Fazzio, T. G. High-resolution chromatin profiling using CUT&RUN. Curr. Protoc. Mol. Biol. 126, 1–22 (2019).
Skene, P. J. & Henikoff, S. A simple method for generating high resolution maps of genome-wide protein binding. eLife 4, 1–9 (2015).
Xu, J. et al. Super-resolution imaging reveals the evolution of higher-order chromatin folding in early carcinogenesis. Nat. Commun. 11, 1–17 (2020).
Schmid, M., Durussel, T. & Laemmli, U. K. ChIC and ChEC: genomic mapping of chromatin proteins. Mol. Cell 16, 147–157 (2004).
Solomon, M. J. & Varshavsky, A. Formaldehyde-mediated DNA-protein crosslinking: a probe for in vivo chromatin structures. Proc. Natl Acad. Sci. USA 82, 6470–6474 (1985).
Gilmour, D. S. & Lis, J. T. In vivo interactions of RNA polymerase II with genes of Drosophila melanogaster. Mol. Cell. Biol. 5, 2009–2018 (1985).
Gilmour, D. S. & Lis, J. T. Detecting protein-DNA interactions in vivo: distribution of RNA polymerase on specific bacterial genes. Proc. Natl Acad. Sci. USA 81, 4275–4279 (1984).
Irvine, R. A., Lin, I. G. & Hsieh, C.-L. DNA methylation has a local effect on transcription and histone acetylation. Mol. Cell. Biol. 22, 6689–6696 (2002).
Albert, I. et al. Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature 446, 572–576 (2007).
Dingwall, C., Lomonossoff, G. P. & Laskey, R. A. High sequence specificity of micrococcal nuclease. Nucleic Acids Res 9, 2659–2674 (1981).
Hörz, W. & Altenburger, W. Sequence specific cleavage of DNA by micrococcal nuclease. Nucleic Acids Res 9, 2643–2658 (1981).
Klein, D. C. & Hainer, S. J. Genomic methods in profiling DNA accessibility and factor localization. Chromosom. Res. 28, 69–85 (2020).
Fosslie, M. et al. Going low to reach high: small-scale ChIP-seq maps new terrain. Wiley Interdiscip. Rev. Syst. Biol. Med. 12, 1–24 (2020).
Buenrostro, J. D., Giresi, P. G., Zaba, L. C., Chang, H. Y. & Greenleaf, W. J. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218 (2013).
Buenrostro, J. D., Wu, B., Chang, H. Y. & Greenleaf, W. J. ATAC-seq: a method for assaying chromatin accessibility genome-wide. Curr. Protoc. Mol. Biol. 2015, 21.29.1–21.29.9 (2015).
Buenrostro, J. D. et al. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature 523, 486–490 (2015).
Schmidl, C., Rendeiro, A. F., Sheffield, N. C. & Bock, C. ChIPmentation: fast, robust, low-input ChIP-seq for histones and transcription factors. Nat. Methods 12, 963–965 (2015).
Cao, Z., Chen, C., He, B., Tan, K. & Lu, C. A microfluidic device for epigenomic profiling using 100 cells. Nat. Methods 12, 959–962 (2015).
Shankaranarayanan, P. et al. Single-tube linear DNA amplification (LinDA) for robust ChIP-seq. Nat. Methods 8, 565–568 (2011).
Rotem, A. et al. Single-cell ChIP-seq reveals cell subpopulations defined by chromatin state. Nat. Biotechnol. 33, 1165–1172 (2015).
Grosselin, K. et al. High-throughput single-cell ChIP-seq identifies heterogeneity of chromatin states in breast cancer. Nat. Genet. 51, 1060–1066 (2019).
Rhee, H. S. & Pugh, B. F. Comprehensive genome-wide protein-DNA interactions detected at single-nucleotide resolution. Cell 147, 1408–1419 (2011).
He, Q., Johnston, J. & Zeitlinger, J. ChIP-nexus enables improved detection of in vivo transcription factor binding footprints. Nat. Biotechnol. 33, 395–401 (2015).
Keller, C. A. et al. Effects of sheared chromatin length on ChIP-seq quality and sensitivity. G3 (Bethesda) https://doi.org/10.1093/g3journal/jkab101 (2021).
Steensel, B. V. & Henikoff, S. Identification of in vivo DNA targets of chromatin proteins using tethered Dam methyltransferase. Nat. Biotechnol. 18, 424–428 (2000).
Kind, J. et al. Genome-wide maps of nuclear lamina interactions in single human. cells. Cell 163, 134–147 (2015).
Zentner, G. E., Kasinathan, S., Xin, B., Rohs, R. & Henikoff, S. ChEC-seq kinetics discriminates transcription factor binding sites by DNA sequence and shape in vivo. Nat. Commun. 6, (2015).
Kaya-Okur, H. S. et al. CUT&Tag for efficient epigenomic profiling of small samples and single cells. Nat. Commun. 10, 1–10 (2019).
Carter, B. et al. Mapping histone modifications in low cell number and single cells using antibody-guided chromatin tagmentation (ACT-seq). Nat. Commun. 10, 1–5 (2019).
Liu, B. et al. The landscape of RNA Pol II binding reveals a stepwise transition during ZGA. Nature 587, 139–144 (2020).
Ku, W. L. et al. Single-cell chromatin immunocleavage sequencing (scChIC-seq) to profile histone modification. Nat. Methods 16, 323–325 (2019).
Skene, P. J. & Henikoff, S. An efficient targeted nuclease strategy for high-resolution mapping of DNA binding sites. eLife 6, 1–35 (2017).
Skene, P. J., Henikoff, J. G. & Henikoff, S. Targeted in situ genome-wide profiling with high efficiency for low cell numbers. Nat. Protoc. 13, 1006–1019 (2018).
Kaya-Okur, H. S., Janssens, D. H., Henikoff, J. G., Ahmad, K. & Henikoff, S. Efficient low-cost chromatin profiling with CUT&Tag. Nat. Protoc. 15, 3264–3283 (2020).
Skene, P. J. & Henikoff, S. A simple method for generating high resolution maps of genome-wide protein binding. eLife 4, 1–9 (2017).
Meers, M. P., Bryson, T. D., Henikoff, J. G. & Henikoff, S. Improved cut&run chromatin profiling tools. eLife 8, 1–16 (2019).
Nagano, T. et al. Single-cell Hi-C reveals cell-to-cell variability in chromosome structure. Nature 502, 59–64 (2013).
Shah, R. N. et al. Examining the roles of H3K4 methylation states with systematically characterized antibodies. Mol. Cell 72, 162–177.e7 (2018).
Zheng, X. Y. & Gehring, M. Low-input chromatin profiling in Arabidopsis endosperm using CUT&RUN. Plant Reprod 32, 63–75 (2019).
Chereji, R. V., Bryson, T. D. & Henikoff, S. Quantitative MNase-seq accurately maps nucleosome occupancy levels. Genome Biol. 20, 198 (2019).
Oomen, M. E., Hansen, A. S., Liu, Y., Darzacq, X. & Dekker, J. CTCF sites display cell cycle-dependent dynamics in factor binding and nucleosome positioning. Genome Res. 29, 236–249 (2019).
Janssens, D. H. et al. Automated in situ chromatin profiling efficiently resolves cell types and gene regulatory programs. Epigenetics Chromatin 11, 1–14 (2018).
Meers, M. P., Tenenbaum, D. & Henikoff, S. Peak calling by Sparse Enrichment Analysis for CUT&RUN chromatin profiling. Epigenetics Chromatin 12, 1–11 (2019).
Zhu, Q., Liu, N., Orkin, S. H. & Yuan, G. C. CUT and RUNTools: a flexible pipeline for CUT and RUN processing and footprint analysis. Genome Biol. 20, 1–12 (2019).
Baek, S. & Lee, I. Single-cell ATAC sequencing analysis: from data preprocessing to hypothesis generation. Comput. Struct. Biotechnol. J. 18, 1429–1439 (2020).
Urrutia, E., Chen, L., Zhou, H. & Jiang, Y. Destin: toolkit for single-cell analysis of chromatin accessibility. Bioinformatics 35, 3818–3820 (2019).
Cai, S., Georgakilas, G. K., Johnson, J. L. & Vahedi, G. A cosine similarity-based method to infer variability of chromatin accessibility at the single-cell level. Front. Genet. 9, 1–10 (2018).
Zamanighomi, M. et al. Unsupervised clustering and epigenetic classification of single cells. Nat. Commun. 9, 1–8 (2018).
Baker, S. M., Rogerson, C., Hayes, A., Sharrocks, A. D. & Rattray, M. Classifying cells with Scasat, a single-cell ATAC-seq analysis tool. Nucleic Acids Res. 47, (2019).
Li, H. et al. The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Ramírez, F., Dündar, F., Diehl, S., Grüning, B. A. & Manke, T. DeepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 42, (2014).
Heinz, S. et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010).
Luo, C. et al. Superovulation strategies for 6 commonly used mouse strains. J. Am. Assoc. Lab. Anim. Sci. 50, 471–478 (2011).
Adli, M. & Bernstein, B. E. Whole-genome chromatin profiling from limited numbers of cells using nano-ChIP-seq. Nat. Protoc. 6, 1656–1668 (2011).
Zwart, W. et al. A carrier-assisted ChIP-seq method for estrogen receptor-chromatin interactions from breast cancer core needle biopsy samples. BMC Genomics 14, (2013).
Valensisi, C., Liao, J. L., Andrus, C., Battle, S. L. & Hawkins, R. D. cChIP-seq: a robust small-scale method for investigation of histone modifications. BMC Genomics 16, 1–11 (2015).
Ng, J. H. et al. In vivo epigenomic profiling of germ cells reveals germ cell molecular signatures. Dev. Cell 24, 324–333 (2013).
Liu, X. et al. Distinct features of H3K4me3 and H3K27me3 chromatin domains in pre-implantation embryos. Nature 537, 558–562 (2016).
Lara-Astiaso, D. et al. Immunogenetics. Chromatin state dynamics during blood formation. Science 345, 943–949 (2014).
van Galen, P. et al. a multiplexed system for quantitative comparisons of chromatin landscapes. Mol. Cell 61, 170–180 (2016).
Zheng, X. et al. Low-cell-number epigenome profiling aids the study of lens aging and hematopoiesis. Cell Rep. 13, 1505–1518 (2015).
Dahl, J. A. et al. Broad histone H3K4me3 domains in mouse oocytes modulate maternal-to-zygotic transition. Nature 537, 548–552 (2016).
Zhang, B. et al. Allelic reprogramming of the histone modification H3K4me3 in early mammalian development. Nature 537, 553–557 (2016).
Zarnegar, M. A., Reinitz, F., Newman, A. M. & Clarke, M. F. Targeted chromatin ligation, a robust epigenetic profiling technique for small cell numbers. Nucleic Acids Res. 45, 1–9 (2017).
Harada, A. et al. A chromatin integration labelling method enables epigenomic profiling with lower input. Nat. Cell Biol. 21, 287–296 (2019).
Akalin, A., Franke, V., Vlahoviček, K., Mason, C. E. & Schübeler, D. Genomation: a toolkit to summarize, annotate and visualize genomic intervals. Bioinformatics 31, 1127–1129 (2015).
Stempor, P. & Ahringer, J. SeqPlots—interactive software for exploratory data analyses, pattern discovery and visualization in genomics. Wellcome Open Res. 1, 14 (2016).
Zhang, Y. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, (2008).
Bailey, T. L. et al. MEME Suite: tools for motif discovery and searching. Nucleic Acids Res. 37, 202–208 (2009).
Chen, X. et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell 133, 1106–1117 (2008).
Acknowledgements
We thank the Henikoff group for the original development of CUT&RUN and discussion regarding application. We thank A. Boskovic and T. Fazzio for assistance in development and application of uliCUT&RUN. We thank members of the Hainer lab for helpful comments on the manuscript. This work was supported by the National Institutes of Health grant R35GM133732 to S.J.H.
Author information
Authors and Affiliations
Contributions
S.J.H. optimized the protocol and performed experiments. B.J.P. analyzed the data with assistance from S.J.H., B.J.P. and S.J.H wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Protocols thanks Sebastian Pott and the other, anonymous reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key references using this protocol
Hainer, S. et al. Cell 177, 1319–1329.e11 (2019): https://doi.org/10.1016/j.cell.2019.03.014
Xu, J. et al. Nat. Commun. 11, 1899 (2020): https://doi.org/10.1038/s41467-020-15718-7
Supplementary information
Supplementary Information
Supplementary Methods and Supplementary Figs. 1 and 2.
Rights and permissions
About this article
Cite this article
Patty, B.J., Hainer, S.J. Transcription factor chromatin profiling genome-wide using uliCUT&RUN in single cells and individual blastocysts. Nat Protoc 16, 2633–2666 (2021). https://doi.org/10.1038/s41596-021-00516-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-021-00516-2
This article is cited by
-
Modeling methyl-sensitive transcription factor motifs with an expanded epigenetic alphabet
Genome Biology (2024)
-
FACT regulates pluripotency through proximal and distal regulation of gene expression in murine embryonic stem cells
BMC Biology (2023)
-
Droplet-based single-cell joint profiling of histone modifications and transcriptomes
Nature Structural & Molecular Biology (2023)
-
A neurodevelopmental epigenetic programme mediated by SMARCD3–DAB1–Reelin signalling is hijacked to promote medulloblastoma metastasis
Nature Cell Biology (2023)
-
Transcriptome and chromatin alterations in social fear indicate association of MEG3 with successful extinction of fear
Molecular Psychiatry (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.