Abstract
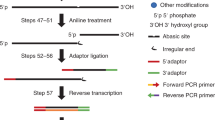
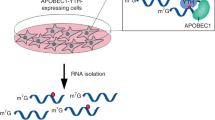
A prerequisite to defining the transcriptome-wide functions of RNA modifications is the ability to accurately determine their location. Here, we present N4-acetylcytidine (ac4C) sequencing (ac4C-seq), a protocol for the quantitative single-nucleotide resolution mapping of cytidine acetylation in RNA. This method exploits the kinetically facile chemical reaction of ac4C with sodium cyanoborohydride under acidic conditions to form a reduced nucleobase. RNA is then fragmented, ligated to an adapter at its 3′ end and reverse transcribed to introduce a non-cognate nucleotide at reduced ac4C sites. After adapter ligation, library preparation and high-throughput sequencing, a bioinformatic pipeline enables identification of ac4C positions on the basis of the presence of C→T misincorporations in reduced samples but not in controls. Unlike antibody-based approaches, ac4C-seq identifies specific ac4C residues and reports on their level of modification. The ac4C-seq library preparation protocol can be completed in ~4 d for transcriptome-wide sequencing.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Code availability
The custom code used for the ‘Calculation of misincorporation level and statistical testing’ section is available at https://github.com/SchwartzLab/ac4c-seq.
References
Zachau, H. G., Dütting, D. & Feldmann, H. Nucleotide sequences of two serine-specific transfer ribonucleic acids. Angew. Chem. Int. Ed. Engl. 5, 422 (1966).
Ohashi, Z. et al. Characterization of C+ located in the first position of the anticodon of Escherichia coli tRNAMet as N4-acetylcytidine. Biochim. Biophys. Acta 262, 209–213 (1972).
Bruenger, E. et al. 5S rRNA modification in the hyperthermophilic archaea Sulfolobus solfataricus and Pyrodictium occultum. FASEB J. 7, 196–200 (1993).
Stern, L. & Schulman, L. H. The role of the minor base N4-acetylcytidine in the function of the Escherichia coli noninitiator methionine transfer RNA. J. Biol. Chem. 253, 6132–6139 (1978).
Sas-Chen, A. et al. Dynamic RNA acetylation revealed by quantitative cross-evolutionary mapping. Nature 583, 638–643 (2020).
Johansson, M. J. & Bystrom, A. S. The Saccharomyces cerevisiae TAN1 gene is required for N4-acetylcytidine formation in tRNA. RNA 10, 712–719 (2004).
Kotelawala, L., Grayhack, E. J. & Phizicky, E. M. Identification of yeast tRNA Um(44) 2′-O-Methyltransferase (Trm44) and demonstration of a Trm44 role in sustaining levels of specific tRNASer species. RNA 14, 158–169 (2008).
Ito, S. et al. A single acetylation of 18 S rRNA is essential for biogenesis of the small ribosomal subunit in Saccharomyces cerevisiae. J. Biol. Chem. 289, 26201–26212 (2014).
Ito, S. et al. Human NAT10 is an ATP-dependent RNA acetyltransferase responsible for N4-acetylcytidine formation in 18 S ribosomal RNA (rRNA). J. Biol. Chem. 289, 35724–35730 (2014).
Sharma, S. et al. Yeast Kre33 and human NAT10 are conserved 18S rRNA cytosine acetyltransferases that modify tRNAs assisted by the adaptor Tan1/THUMPD1. Nucleic Acids Res. 43, 2242–2258 (2015).
Sharma, S. et al. Specialized box C/D snoRNPs act as antisense guides to target RNA base acetylation. PLoS Genet. 13, e1006804 (2017).
Thomas, J. M., Bryson, K. M. & Meier, J. L. Nucleotide resolution sequencing of N4-acetylcytidine in RNA. Methods Enzymol. 621, 31–51 (2019).
Wang, T. et al. Identification and characterization of essential genes in the human genome. Science 350, 1096–1101 (2015).
Dempster, J. M. et al. Extracting biological insights from the Project Achilles genome-scale CRISPR screens in cancer cell lines. Preprint at bioRxiv https://doi.org/10.1101/720243 (2019).
Larrieu, D., Britton, S., Demir, M., Rodriguez, R. & Jackson, S. P. Chemical inhibition of NAT10 corrects defects of laminopathic cells. Science 344, 527–532 (2014).
Tschida, B. R. et al. Sleeping Beauty insertional mutagenesis in mice identifies drivers of steatosis-associated hepatic tumors. Cancer Res. 77, 6576–6588 (2017).
Ryvkin, P. et al. HAMR: high-throughput annotation of modified ribonucleotides. RNA 19, 1684–1692 (2013).
Cerutti, P. & Miller, N. Selective reduction of yeast transfer ribonucleic acid with sodium borohydride. J. Mol. Biol. 67, 90260–90264 (1967).
Thomas, J. M. et al. A chemical signature for cytidine acetylation in RNA. J. Am. Chem. Soc. 140, 12667–12670 (2018).
Sinclair, W. R. et al. Profiling cytidine acetylation with specific affinity and reactivity. ACS Chem. Biol. 12, 2922–2926 (2017).
Tsai, K. et al. Acetylation of cytidine residues boosts HIV-1 gene expression by increasing viral RNA stability. Cell Host Microbe 28, 306–312.e6 (2020).
Balmus, G. et al. Targeting of NAT10 enhances healthspan in a mouse model of human accelerated aging syndrome. Nat. Commun. 9, 1700 (2018).
Li, Q. et al. NAT10 is upregulated in hepatocellular carcinoma and enhances mutant p53 activity. BMC Cancer 17, 605 (2017).
Liu, X. et al. Deacetylation of NAT10 by Sirt1 promotes the transition from rRNA biogenesis to autophagy upon energy stress. Nucleic Acids Res. 46, 9601–9616 (2018).
Lin, S. et al. Mettl1/Wdr4-mediated mG tRNA methylome is required for normal mRNA translation and embryonic stem cell self-renewal and differentiation. Mol. Cell 71, 244–255.e5 (2018).
Marchand, V. et al. AlkAniline-Seq: profiling of M7 G and M3 C RNA modifications at single nucleotide resolution. Angew. Chem. Int. Ed. Engl. 57, 16785–16790 (2018).
Ikeuchi, Y., Kitahara, K. & Suzuki, T. The RNA acetyltransferase driven by ATP hydrolysis synthesizes N4-acetylcytidine of tRNA anticodon. EMBO J. 27, 2194–2203 (2008).
Arango, D. et al. Acetylation of cytidine in mRNA promotes translation efficiency. Cell 175, 1872–1886.e24 (2018).
Safra, M. et al. The m1A landscape on cytosolic and mitochondrial mRNA at single-base resolution. Nature 551, 251–255 (2017).
Grozhik, A. V. et al. Antibody cross-reactivity accounts for widespread appearance of m1A in 5′UTRs. Nat. Commun. 10, 5126 (2019).
Helm, M., Lyko, F. & Motorin, Y. Limited antibody specificity compromises epitranscriptomic analyses. Nat. Commun. 10, 5669 (2019).
Liu, H. et al. Accurate detection of m6A RNA modifications in native RNA sequences. Nat. Commun. 10, 4079 (2019).
Grünberger, F. et al. Exploring prokaryotic transcription, operon structures, rRNA maturation and modifications using nanopore-based native RNA sequencing. Preprint at bioRxiv https://doi.org/10.1101/2019.12.18.880849 (2019).
Sas-Chen, A. & Schwartz, S. Misincorporation signatures for detecting modifications in mRNA: not as simple as it sounds. Methods 156, 53–59 (2019).
Orita, I. et al. Random mutagenesis of a hyperthermophilic archaeon identified tRNA modifications associated with cellular hyperthermotolerance. Nucleic Acids Res. 47, 1964–1976 (2019).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. J. Bioinform. 29, 15–21 (2013).
Piechotta, M. et al. JACUSA: site-specific identification of RNA editing events from replicate sequencing data. BMC Bioinformatics 18, 7 (2017).
Sexton, A. N., Wang, P. Y., Rutenberg-Schoenberg, M. & Simon, M. D. Interpreting reverse transcriptase termination and mutation events for greater insight into the chemical probing of RNA. Biochemistry 56, 4713–4721 (2017).
Cozen, A. E. et al. ARM-Seq: AlkB-facilitated RNA methylation sequencing reveals a complex landscape of modified tRNA fragments. Nat. Methods 12, 879–884 (2015).
Zheng, G. et al. Efficient and quantitative high-throughput tRNA sequencing. Nat. Methods 12, 835–837 (2015).
Quinodoz, S. A. et al. Higher-order inter-chromosomal hubs shape 3D genome organization in the nucleus. Cell 174, 744–757 (2018).
Li, H. et al. The Sequence Alignment/Map format and SAMtools. J. Bioinform 25, 2078–2079 (2009).
R Core Team. R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, Vienna, Austria, 2020) Available at https://www.R-project.org/
Bates, D., Mächler, M., Bolker, B. & Walker, S. Fitting linear mixed-effects models using lme4. J. Stat. Softw. 67, 1–48 (2015).
Chomczynski, P. A reagent for the single-step simultaneous isolation of RNA, DNA and proteins from cell and tissue samples. Biotechniques 15, 532–534 (1993).
Hummon, A. B., Lim, S. R., Difilippantonio, M. J. & Ried, T. Isolation and solubilization of proteins after TRIzol® extraction of RNA and DNA from patient material following prolonged storage. Biotechniques 42, 467–472 (2007).
Collart, M. A. & Oliviero, S. Preparation of yeast RNA. Curr. Protoc. Mol. Biol. Ch. 13, Unit 13.12 (2001).
Robinson, J. et al. Integrative genomics viewer. Nat. Biotechnol. 29, 24–26 (2011).
Acknowledgements
We thank members of the Schwartz and Meier laboratories for many helpful comments. S.S. is funded by the Israel Science Foundation (543165), the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (grant agreement no. 714023) and the Estate of Emile Mimran. S.S. is the incumbent of the Robert Edward and Roselyn Rich Manson Career Development Chair in Perpetuity. J.L.M. is supported by the Intramural Research Program of the National Institutes of Health (NIH), the National Cancer Institute, The Center for Cancer Research (ZIA BC011488-06).
Author information
Authors and Affiliations
Contributions
A.S.-C., S.T.G., J.L.M. and S.S. developed the protocol. A.S.-C. and S.T.G. performed the experiments. A.S.-C. designed the bioinformatic pipeline and analyzed the data. S.T.G. A.S.-C., S.S. and J.L.M. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Protocols thanks the anonymous reviewers for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key references using this protocol
Sinclair, W. et al. ACS Chem. Biol. 12, 2922–2926 (2017): https://doi.org/10.1021/acschembio.7b00734
Thomas, J. et al. J. Am. Chem. Soc. 140, 12667–12670 (2018): https://doi.org/10.1021/jacs.8b06636
Sas-Chen, A. et al. Nature 583, 638–643 (2020): https://doi.org/10.1038/s41586-020-2418-2
Rights and permissions
About this article
Cite this article
Thalalla Gamage, S., Sas-Chen, A., Schwartz, S. et al. Quantitative nucleotide resolution profiling of RNA cytidine acetylation by ac4C-seq. Nat Protoc 16, 2286–2307 (2021). https://doi.org/10.1038/s41596-021-00501-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-021-00501-9
This article is cited by
-
Recent advances in the potential role of RNA N4-acetylcytidine in cancer progression
Cell Communication and Signaling (2024)
-
RNA modifications in cellular metabolism: implications for metabolism-targeted therapy and immunotherapy
Signal Transduction and Targeted Therapy (2024)
-
Dissecting the oncogenic properties of essential RNA-modifying enzymes: a focus on NAT10
Oncogene (2024)
-
RNPS1 stabilizes NAT10 protein to facilitate translation in cancer via tRNA ac4C modification
International Journal of Oral Science (2024)
-
RNA modification: mechanisms and therapeutic targets
Molecular Biomedicine (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.