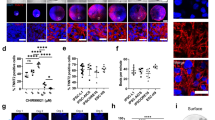
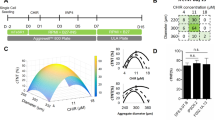
Abstract
Tissue-like structures from human pluripotent stem cells containing multiple cell types are transforming our ability to model and understand human development and disease. Here we describe a protocol to generate cardiomyocytes (CMs), cardiac fibroblasts (CFs) and cardiac endothelial cells (ECs), the three principal cell types in the heart, from human induced pluripotent stem cells (hiPSCs) and combine them in three-dimensional (3D) cardiac microtissues (MTs). We include details of how to differentiate, isolate, cryopreserve and thaw the component cells and how to construct and analyze the MTs. The protocol supports hiPSC-CM maturation and allows replacement of one or more of the three heart cell types in the MTs with isogenic variants bearing disease mutations. Differentiation of each cell type takes ~30 d, while MT formation and maturation requires another 20 d. No specialist equipment is needed and the method is inexpensive, requiring just 5,000 cells per MT.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout









Similar content being viewed by others
Data availability
The datasets generated and/or analyzed during the current study are available in Giacomelli et al. 202011 and from the corresponding authors upon request.
Code availability
MUSCLEMOTION is freely accessible on the web at https://gitlab.com/bjvanmeer/MUSCLEMOTION or https://github.com/l-sala/MUSCLEMOTION/. The license is a GNU GENERAL PUBLIC LICENSE version 3 (GPL v3), which can be found at https://github.com/l-sala/MUSCLEMOTION/blob/master/LICENSE. After downloading the macro, users can access the code through a text editor or directly through FIJI. When using MUSCLEMOTION, users are encouraged to read Sala, L. et al.61 (https://www.ahajournals.org/doi/suppl/10.1161/CIRCRESAHA.117.312067).
References
Broutier, L. et al. Culture and establishment of self-renewing human and mouse adult liver and pancreas 3D organoids and their genetic manipulation. Nat. Protoc. 11, 1724–1743 (2016).
Koike, H. et al. Modelling human hepato-biliary-pancreatic organogenesis from the foregut-midgut boundary. Nature 574, 112–116 (2019).
Sato, T. et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology 141, 1762–1772 (2011).
Tirziu, D., Giordano, F. J. & Simons, M. Cell communications in the heart. Circulation 122, 928–937 (2010).
Bergmann, O. et al. Dynamics of cell generation and turnover in the human heart. Cell 161, 1566–1575 (2015).
Zhou, P. & Pu, W. T. Recounting cardiac cellular composition. Circ. Res. 118, 368–370 (2016).
Pinto, A. R. et al. Revisiting cardiac cellular composition. Circ. Res. 118, 400–409 (2016).
Banerjee, I., Fuseler, J. W., Price, R. L., Borg, T. K. & Baudino, T. A. Determination of cell types and numbers during cardiac development in the neonatal and adult rat and mouse. Am. J. Physiol. Heart Circ. Physiol. 293, H1883–H1891 (2007).
Furtado, M. B., Nim, H. T., Boyd, S. E. & Rosenthal, N. A. View from the heart: cardiac fibroblasts in development, scarring and regeneration. Development 143, 387–3971 (2016).
Brutsaert, D. L. Cardiac endothelial-myocardial signaling: its role in cardiac growth, contractile performance, and rhythmicity. Physiol. Rev. 83, 59–1151 (2003).
Giacomelli, E. et al. Human-iPSC-derived cardiac stromal cells enhance maturation in 3D cardiac microtissues and reveal non-cardiomyocyte contributions to heart disease. Cell Stem Cell 26, 862–879 e811 (2020).
Garry, D. J. & Olson, E. N. A common progenitor at the heart of development. Cell 127, 1101–1104 (2006).
Moretti, A. et al. Multipotent embryonic isl1+ progenitor cells lead to cardiac, smooth muscle, and endothelial cell diversification. Cell 127, 1151–1165 (2006).
van Wijk, B. & van den Hoff, M. Epicardium and myocardium originate from a common cardiogenic precursor pool. Trends Cardiovasc. Med. 20, 1–7 (2010).
Giacomelli, E. et al. Three-dimensional cardiac microtissues composed of cardiomyocytes and endothelial cells co-differentiated from human pluripotent stem cells. Development 144, 1008–1017 (2017).
Iwamiya, T., Matsuura, K., Masuda, S., Shimizu, T. & Okano, T. Cardiac fibroblast-derived VCAM-1 enhances cardiomyocyte proliferation for fabrication of bioengineered cardiac tissue. Regen. Ther. 4, 92–102 (2016).
Guadix, J. A. et al. Human pluripotent stem cell differentiation into functional epicardial progenitor cells. Stem Cell Reports 9, 1754–1764 (2017).
Tallquist, M. D. & Molkentin, J. D. Redefining the identity of cardiac fibroblasts. Nat. Rev. Cardiol 14, 484–491 (2017).
Zhao, J. et al. Efficient differentiation of TBX18(+)/WT1(+) epicardial-like cells from human pluripotent stem cells using small molecular compounds. Stem Cells Dev. 26, 528–540 (2017).
Bich, L., Pradeu, T. & Moreau, J. F. Understanding multicellularity: the functional organization of the intercellular space. Front. Physiol. 10, 1170 (2019).
Wan, A. C. A. Recapitulating cell-cell interactions for organoid construction—are biomaterials dispensable? Trends Biotechnol. 34, 711–721 (2016).
Mills, R. J. et al. Functional screening in human cardiac organoids reveals a metabolic mechanism for cardiomyocyte cell cycle arrest. Proc. Natl Acad. Sci. USA 114, E8372–E8381 (2017).
Nunes, S. S. et al. Biowire: a platform for maturation of human pluripotent stem cell-derived cardiomyocytes. Nat. Methods 10, 781–787 (2013).
Gao, L. et al. Myocardial tissue engineering with cells derived from human-induced pluripotent stem cells and a native-like, high-resolution, 3-dimensionally printed scaffold. Circ. Res. 120, 1318–1325 (2017).
Ong, C. S. et al. Biomaterial-free three-dimensional bioprinting of cardiac tissue using human induced pluripotent stem cell derived cardiomyocytes. Sci. Rep. 7, 4566 (2017).
Goldfracht, I. et al. Engineered heart tissue models from hiPSC-derived cardiomyocytes and cardiac ECM for disease modeling and drug testing applications. Acta Biomater. 92, 145–159 (2019).
Wang, G. et al. Modeling the mitochondrial cardiomyopathy of Barth syndrome with induced pluripotent stem cell and heart-on-chip technologies. Nat. Med. 20, 616–623 (2014).
Ma, Z. et al. Contractile deficits in engineered cardiac microtissues as a result of MYBPC3 deficiency and mechanical overload. Nat. Biomed. Eng. 2, 955–967 (2018).
Wanjare, M. et al. Vascularization of engineered spatially patterned myocardial tissue derived from human pluripotent stem cells in vivo. Front. Bioeng. Biotechnol. 7, 208 (2019).
Zhao, Y. et al. A platform for generation of chamber-specific cardiac tissues and disease modeling. Cell 176, 913–927 e918 (2019).
Tulloch, N. L. et al. Growth of engineered human myocardium with mechanical loading and vascular coculture. Circ. Res. 109, 47–59 (2011).
Schaaf, S. et al. Human engineered heart tissue as a versatile tool in basic research and preclinical toxicology. PLoS ONE 6, e26397 (2011).
Burridge, P. W. et al. Multi-cellular interactions sustain long-term contractility of human pluripotent stem cell-derived cardiomyocytes. Am. J. Transl. Res. 6, 724–735 (2014).
Hinson, J. T. et al. Heart disease. Titin mutations in iPS cells define sarcomere insufficiency as a cause of dilated cardiomyopathy. Science 349, 982–986 (2015).
Mannhardt, I. et al. Human engineered heart tissue: analysis of contractile force. Stem Cell Reports 7, 29–42 (2016).
Keung, W. et al. Non-cell autonomous cues for enhanced functionality of human embryonic stem cell-derived cardiomyocytes via maturation of sarcolemmal and mitochondrial KATP channels. Sci. Rep. 6, 34154 (2016).
Tiburcy, M. et al. Defined engineered human myocardium with advanced maturation for applications in heart failure modeling and repair. Circulation 135, 1832–1847 (2017).
Lee, E. K. et al. Machine learning of human pluripotent stem cell-derived engineered cardiac tissue contractility for automated drug classification. Stem Cell Reports 9, 1560–1572 (2017).
Shadrin, I. Y. et al. Cardiopatch platform enables maturation and scale-up of human pluripotent stem cell-derived engineered heart tissues. Nat. Commun. 8, 1825 (2017).
Ronaldson-Bouchard, K. et al. Advanced maturation of human cardiac tissue grown from pluripotent stem cells. Nature 556, 239–243 (2018).
Tsuruyama, S., Matsuura, K., Sakaguchi, K. & Shimizu, T. Pulsatile tubular cardiac tissues fabricated by wrapping human iPS cells-derived cardiomyocyte sheets. Regen. Ther. 11, 297–305 (2019).
Mills, R. J. et al. Drug screening in human PSC-cardiac organoids identifies pro-proliferative compounds acting via the mevalonate pathway. Cell Stem Cell 24, 895–907 e896 (2019).
Goldfracht, I. et al. Generating ring-shaped engineered heart tissues from ventricular and atrial human pluripotent stem cell-derived cardiomyocytes. Nat. Commun. 11, 75 (2020).
Jang, Y. et al. Modulating cardiomyocyte and fibroblast interaction using layer-by-layer deposition facilitates synchronisation of cardiac macro tissues. Soft Matter 16, 428–434 (2020).
Dostanic, M. et al. A miniaturized EHT platform for accurate measurements of tissue contractile properties. J. of Microelectromech. Syst. 29, 881–887 (2020).
Masumoto, H. et al. The myocardial regenerative potential of three-dimensional engineered cardiac tissues composed of multiple human iPS cell-derived cardiovascular cell lineages. Sci. Rep. 6, 29933 (2016).
Huebsch, N. et al. Miniaturized iPS-cell-derived cardiac muscles for physiologically relevant drug response analyses. Sci. Rep. 6, 24726 (2016).
Zimmermann, W. H. et al. Tissue engineering of a differentiated cardiac muscle construct. Circ. Res. 90, 223–230 (2002).
Ravenscroft, S. M., Pointon, A., Williams, A. W., Cross, M. J. & Sidaway, J. E. Cardiac non-myocyte cells show enhanced pharmacological function suggestive of contractile maturity in stem cell derived cardiomyocyte microtissues. Toxicol. Sci. 152, 99–112 (2016).
Pointon, A. et al. From the cover: high-throughput imaging of cardiac microtissues for the assessment of cardiac contraction during drug discovery. Toxicol. Sci. 155, 444–457 (2017).
Forsythe, S. D. et al. Environmental toxin screening using human-derived 3D Bioengineered liver and cardiac organoids. Front. Public Health 6, 103 (2018).
Stevens, K. R., Pabon, L., Muskheli, V. & Murry, C. E. Scaffold-free human cardiac tissue patch created from embryonic stem cells. Tissue Eng. Part A 15, 1211–1222 (2009).
Polonchuk, L. et al. Cardiac spheroids as promising in vitro models to study the human heart microenvironment. Sci. Rep. 7, 7005 (2017).
Sebastiao, M. J. et al. Bioreactor-based 3D human myocardial ischemia/reperfusion in vitro model: a novel tool to unveil key paracrine factors upon acute myocardial infarction. Transl. Res. 215, 57–74 (2020).
Richards, D. J. et al. Human cardiac organoids for the modelling of myocardial infarction and drug cardiotoxicity. Nat. Biomed. Eng. 4, 446–462 (2020).
Qiao, X. et al. Uncoupling DNA damage from chromatin damage to detoxify doxorubicin. Proc. Natl Acad. Sci. USA 117, 15182–15192 (2020).
Veerman, C. C. et al. Immaturity of human stem-cell-derived cardiomyocytes in culture: fatal flaw or soluble problem? Stem Cells Dev. 24, 1035–1052 (2015).
Gaudesius, G., Miragoli, M., Thomas, S. P. & Rohr, S. Coupling of cardiac electrical activity over extended distances by fibroblasts of cardiac origin. Circ. Res. 93, 421–428 (2003).
De Simone, S. A. et al. The role of membrane capacitance in cardiac impulse conduction: an optogenetic study with non-excitable cells coupled to cardiomyocytes. Front. Physiol. 11, 194 (2020).
Quaife-Ryan, G. A. et al. Multicellular transcriptional analysis of mammalian heart regeneration. Circulation 136, 1123–1139 (2017).
Sala, L. et al. MUSCLEMOTION: a versatile open software tool to quantify cardiomyocyte and cardiac muscle contraction in vitro and in vivo. Circ. Res. 122, e5–e16 (2018).
Mastikhina, O. et al. Human cardiac fibrosis-on-a-chip model recapitulates disease hallmarks and can serve as a platform for drug testing. Biomaterials 233, 119741 (2020).
van den Berg, C. W., Elliott, D. A., Braam, S. R., Mummery, C. L. & Davis, R. P. Differentiation of human pluripotent stem cells to cardiomyocytes under defined conditions. Methods Mol. Biol. 1353, 163–180 (2016).
Sala, L., Ward-van Oostwaard, D., Tertoolen, L. G. J., Mummery, C. L. & Bellin, M. Electrophysiological analysis of human pluripotent stem cell-derived cardiomyocytes (hPSC-CMs) using multi-electrode arrays (MEAs). J. Vis. Exp. https://doi.org/10.3791/55587 (2017).
Giacomelli, E., Bellin, M., Orlova, V. V. & Mummery, C. L. Co-differentiation of human pluripotent stem cells-derived cardiomyocytes and endothelial cells from cardiac mesoderm provides a three-dimensional model of cardiac microtissue. Curr. Protoc. Hum. Genet. 95, 21.9.1–21.9.22 (2017).
van den Brink, L. et al. Cryopreservation of human pluripotent stem cell-derived cardiomyocytes is not detrimental to their molecular and functional properties. Stem Cell Res. 43, 101698 (2020).
Burridge, P. W. et al. Chemically defined generation of human cardiomyocytes. Nat. Methods 11, 855–860 (2014).
Lian, X. et al. Directed cardiomyocyte differentiation from human pluripotent stem cells by modulating Wnt/beta-catenin signaling under fully defined conditions. Nat. Protoc. 8, 162–175 (2013).
Tohyama, S. et al. Distinct metabolic flow enables large-scale purification of mouse and human pluripotent stem cell-derived cardiomyocytes. Cell Stem Cell 12, 127–137 (2013).
Moretti, A. et al. Patient-specific induced pluripotent stem-cell models for long-QT syndrome. N. Engl. J. Med. 363, 1397–1409 (2010).
Zhang, M. et al. Recessive cardiac phenotypes in induced pluripotent stem cell models of Jervell and Lange-Nielsen syndrome: disease mechanisms and pharmacological rescue. Proc. Natl Acad. Sci. USA 111, E5383–E5392 (2014).
Meraviglia, V. et al. Generation of human induced pluripotent stem cell line LUMCi027-A and its isogenic gene-corrected line from a patient affected by arrhythmogenic cardiomyopathy and carrying the c.2013delC PKP2 mutation. Stem Cell Res. 46, 101835 (2020).
Assou, S., Bouckenheimer, J. & De Vos, J. Concise review: assessing the genome integrity of human induced pluripotent stem cells: what quality control metrics? Stem Cells 36, 814–821 (2018).
Stacey, G. N. et al. Stem cell culture conditions and stability: a joint workshop of the PluriMes Consortium and Pluripotent Stem Cell Platform. Regen. Med. 14, 243–255 (2019).
Miller, L. R. et al. Considering sex as a biological variable in preclinical research. FASEB J. 31, 29–34 (2017).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Pioner, J. M. et al. Isolation and mechanical measurements of myofibrils from human induced pluripotent stem cell-derived cardiomyocytes. Stem Cell Reports 6, 885–896 (2016).
Ferrantini, C. et al. Impact of detubulation on force and kinetics of cardiac muscle contraction. J. Gen. Physiol. 143, 783–797 (2014).
van Meer, B. J. et al. Quantification of muscle contraction in vitro and in vivo using MUSCLEMOTION software: from stem cell-derived cardiomyocytes to zebrafish and human hearts. Curr. Protoc. Hum. Genet. 99, e67 (2018).
Acknowledgements
We thank D. Ward-van Oostwaard, M. P. H. Mol, L. G. J. Tertoolen, A. Krotenberg Garcia and A. Cochrane for technical assistance. This project was supported by European Research Council (ERCAdG 323182 STEMCARDIOVASC; ERCStG 638030 STEMCARDIORISK); European Community’s Seventh Framework Programme (FP7/2007-2013) under grant agreement No. 602423; European Union’s Horizon 2020 research and innovation Programme (H2020 Technobeat) under grant agreement No. 668724; The Netherlands Organ-on-Chip Initiative is an NWO Gravitation project (024.003.001) funded by the Ministry of Education, Culture and Science of the government of the Netherlands; Transnational Research Project on Cardiovascular Diseases (JTC2016_FP-40-021 ACM-HF); The Netherlands Organisation for Health Research and Development (ZonMW) (MKMD project No. 114022504; VIDI project No. 91715303; TAS project No. 446002501); Health~Holland TKI-LSH PPP-allowance (LSHM17013-H007); Individual Fellowship under the Marie Sklodowska Curie Grant Agreement No. 838985.
Author information
Authors and Affiliations
Contributions
G.C. and V.M. designed and performed the experiments, analyzed the data and wrote the manuscript. E.G developed and optimized the cardiac MT protocol, acquired and analyzed the data and wrote the manuscript. R.v.H. and L.Y acquired and analyzed the data and wrote the manuscript. R.P.D. contributed to drafting the manuscript and revising it for important intellectual content. M.B., V.O. and C.L.M. conceived, designed and supervised the protocol, acquired the funding and wrote and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
C.L.M. is co-founder of Pluriomics (now Ncardia) bv.
Additional information
Peer review information Nature Protocols thanks Sara Nunes de Vasconcelos, Wolfram H Zimmermann and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key references using this protocol
Giacomelli, E. et al. Cell Stem Cell 26, 862–879 e811 (2020): https://doi.org/10.1016/j.stem.2020.05.004
Giacomelli E. et al. Curr. Protoc. Hum. Genet. 95, 21.9.1–21.9.22 (2017): https://doi.org/10.1002/cphg.46
Giacomelli, E. et al. Development 144, 1008–1017 (2017): https://doi.org/10.1242/dev.143438
Supplementary information
Supplementary Information
Supplementary Figs. 1–3.
Supplementary Table 1
Raw data for results summarized in Table 3. Table contains morphological, contraction and single-cell electrophysiological parameters measured in LUMC0099iCTRL04 and LUMC0020iCTRL06 hiPSC lines.
Supplementary Video 1
Beating monolayer hiPSC-CMs after 21 d of differentiation using LI-BPEL protocol with lactate purification
Supplementary Video 2
Beating monolayer hiPSC-CMs after 21 d of differentiation using mBPEL protocol
Supplementary Video 3
Time-lapse movie of the first 72 h of MT formation (time frame: 15 min)
Supplementary Video 4
Movie of representative MT showing CD31 positive ECs organized in a vessel-like structure (in red) and CMs stained with ACTN2 (in green) (15 f.p.s.)
Supplementary Video 5
Contracting MT paced at 1 Hz at after 21 d in culture
Supplementary Video 6
Contracting MT loaded with Fluo-4, paced at 1.5 Hz after 21 d in culture
Rights and permissions
About this article
Cite this article
Campostrini, G., Meraviglia, V., Giacomelli, E. et al. Generation, functional analysis and applications of isogenic three-dimensional self-aggregating cardiac microtissues from human pluripotent stem cells. Nat Protoc 16, 2213–2256 (2021). https://doi.org/10.1038/s41596-021-00497-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-021-00497-2
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.