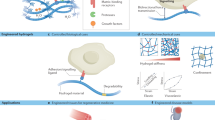
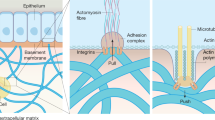
Abstract
Growing interest in exploring mechanically mediated biological phenomena has resulted in cell culture substrates and 3D matrices with variable stiffnesses becoming standard tools in biology labs. However, correlating stiffness with biological outcomes and comparing results between research groups is hampered by variability in the methods used to determine Young’s (elastic) modulus, E, and by the inaccessibility of relevant mechanical engineering protocols to most biology labs. Here, we describe a protocol for measuring E of soft 2D surfaces and 3D hydrogels using atomic force microscopy (AFM) force spectroscopy. We provide instructions for preparing hydrogels with and without encapsulated live cells, and provide a method for mounting samples within the AFM. We also provide details on how to calibrate the instrument, and give step-by-step instructions for collecting force-displacement curves in both manual and automatic modes (stiffness mapping). We then provide details on how to apply either the Hertz or the Oliver-Pharr model to calculate E, and give additional instructions to aid the user in plotting data distributions and carrying out statistical analyses. We also provide instructions for inferring differential matrix remodeling activity in hydrogels containing encapsulated single cells or organoids. Our protocol is suitable for probing a range of synthetic and naturally derived polymeric hydrogels such as polyethylene glycol, polyacrylamide, hyaluronic acid, collagen, or Matrigel. Although sample preparation timings will vary, a user with introductory training to AFM will be able to use this protocol to characterize the mechanical properties of two to six soft surfaces or 3D hydrogels in a single day.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Code availability
The MATLAB code described in this paper is freely available at https://github.com/eileengentleman/AFM-code.
References
Evans, N. D. & Gentleman, E. The role of material structure and mechanical properties in cell-matrix interactions. J. Mater. Chem. B 2, 2345–2356 (2014).
Engler, A. J., Sen, S., Sweeney, H. L. & Discher, D. E. Matrix elasticity directs stem cell lineage specification. Cell 126, 677–689 (2006).
Evans, N. D. et al. Substrate stiffness affects early differentiation events in embryonic stem cells. Eur. Cell. Mater. 18, 1–14 (2009).
Krishnan, R. et al. Substrate stiffening promotes endothelial monolayer disruption through enhanced physical forces. Am. J. Physiol. Cell Physiol. 300, C146–C154 (2011).
Mammoto, A., Mammoto, T. & Ingber, D. E. Mechanosensitive mechanisms in transcriptional regulation. J. Cell Sci. 125, 3061–3073 (2012).
Engler, A. J. et al. Myotubes differentiate optimally on substrates with tissue-like stiffness: pathological implications for soft or stiff microenvironments. J. Cell. Biol. 166, 877–887 (2004).
Wells, R. G. The role of matrix stiffness in regulating cell behavior. Hepatology 47, 1394–1400 (2008).
Levental, K. R. et al. Matrix crosslinking forces tumor progression by enhancing integrin signaling. Cell 139, 891–906 (2009).
Martin, L. J. & Boyd, N. F. Mammographic density. Potential mechanisms of breast cancer risk associated with mammographic density: hypotheses based on epidemiological evidence. Breast Cancer Res. 10, 201–215 (2008).
Wei, S. C. et al. Matrix stiffness drives epithelial-mesenchymal transition and tumour metastasis through a TWIST1-G3BP2 mechanotransduction pathway. Nat. Cell Biol. 17, 678 (2015).
Baumgart, F. Stiffness - an unknown world of mechanical science? Injury 31, 14–84 (2000).
Krieg, M. et al. Atomic force microscopy-based mechanobiology. Nat. Rev. Phys. 1, 41–57 (2019).
Thompson, D. W. On Growth and Form 1942 edn (Cambridge University Press, 1917).
Emerman, J. T. & Pitelka, D. R. Maintenance and induction of morphological differentiation in dissociated mammary epithelium on floating collagen membranes. In Vitro 13, 316–328 (1977).
Pelham, R. J. & Wang, Y. Cell locomotion and focal adhesions are regulated by substrate flexibility. Proc. Natl Acad. Sci. USA 94, 13661–13665 (1997).
Discher, D. E., Janmey, P. & Wang, Y. L. Tissue cells feel and respond to the stiffness of their substrate. Science 310, 1139–1143 (2005).
Foyt, D. A. et al. Hypoxia impacts human MSC response to substrate stiffness during chondrogenic differentiation. Acta Biomater. 15, 73–83 (2019).
Chin, M. H. W., Norman, M. D. A., Gentleman, E., Coppens, M.-O. & Day, R. M. A hydrogel-integrated culture device to interrogate T cell activation with physicochemical cues. ACS Appl. Mater. Interfaces 12, 47355–47367 (2020).
Huebsch, N. et al. Harnessing traction-mediated manipulation of the cell/matrix interface to control stem-cell fate. Nat. Mater. 9, 518–526 (2010).
Khetan, S. et al. Degradation-mediated cellular traction directs stem cell fate in covalently crosslinked three-dimensional hydrogels. Nat. Mater. 12, 458–465 (2013).
Blache, U., Stevens, M. M. & Gentleman, E. Harnessing the secreted matrix to engineer tissues. Nat. Biomed. Eng. 4, 357–363 (2020).
Ferreira, S. A. et al. Bi-directional cell-pericellular matrix interactions direct stem cell fate. Nat. Commun. 9, 4049 (2018).
Loebel, C., Mauck, R. L. & Burdick, J. A. Local nascent protein deposition and remodelling guide mesenchymal stromal cell mechanosensing and fate in three-dimensional hydrogels. Nat. Mater. 18, 883–891 (2019).
Jowett, G. M. et al. ILC1 drive intestinal epithelial and matrix remodelling. Nat. Mater. 20, 250–259 (2021).
Barriga, E. H., Franze, K., Charras, G. & Mayor, R. Tissue stiffening coordinates morphogenesis by triggering collective cell migration in vivo. Nature 554, 523–527 (2018).
Gilbert, P. M. et al. Substrate elasticity regulates skeletal muscle stem cell self-renewal in culture. Science 329, 1078–1081 (2010).
McKee, C. T., Last, J. A., Russell, P. & Murphy, C. J. Indentation versus tensile measurements of Young’s modulus for soft biological tissues. Tissue Eng. Part B Rev. 17, 155–164 (2011).
Denisin, A. K. & Pruitt, B. L. Tuning the range of polyacrylamide gel stiffness for mechanobiology applications. ACS Appl. Mater. Interfaces 8, 21893–21902 (2016).
Prager-Khoutorsky, M. et al. Fibroblast polarization is a matrix-rigidity-dependent process controlled by focal adhesion mechanosensing. Nat. Cell Biol. 13, 1457–1465 (2011).
Trappmann, B. et al. Extracellular-matrix tethering regulates stem-cell fate. Nat. Mater. 11, 642–649 (2012).
Wen, J. H. et al. Interplay of matrix stiffness and protein tethering in stem cell differentiation. Nat. Mater. 13, 979–987 (2014).
Oyen, M. L. Nanoindentation of biological and biomimetic materials. Exp. Tech. 37, 73–87 (2013).
Megone, W., Roohpour, N. & Gautrot, J. E. Impact of surface adhesion and sample heterogeneity on the multiscale mechanical characterisation of soft biomaterials. Sci. Rep. 8, 6780 (2018).
Schultz, K. M. & Furst, E. M. Microrheology of biomaterial hydrogelators. Soft Matter 8, 6198–6205 (2012).
Ziemann, F., Radler, J. & Sackmann, E. Local measurements of viscoelastic moduli of entangled actin networks using an oscillating magnetic bead micro-rheometer. Biophys. J. 66, 2210–2216 (1994).
Campas, O. et al. Quantifying cell-generated mechanical forces within living embryonic tissues. Nat. Methods 11, 183–189 (2014).
Wang, S. & Larin, K. V. Optical coherence elastography for tissue characterization: a review. J. Biophotonics 8, 279–302 (2015).
Scarcelli, G. & Yun, S. H. Confocal Brillouin microscopy for three-dimensional mechanical imaging. Nat. Photonics 2, 39–43 (2007).
Shi, Y., Glaser, K. J., Venkatesh, S. K., Ben-Abraham, E. I. & Ehman, R. L. Feasibility of using 3D MR elastography to determine pancreatic stiffness in healthy volunteers. J. Magn. Reson. Imaging 41, 369–375 (2015).
Anvari, A., Dhyani, M., Stephen, A. E. & Samir, A. E. Reliability of shear-wave elastography estimates of the young modulus of tissue in follicular thyroid neoplasms. Am. J. Roentgenol. 206, 609–616 (2016).
Schultz, K. M., Kyburz, K. A. & Anseth, K. S. Measuring dynamic cell-material interactions and remodeling during 3D human mesenchymal stem cell migration in hydrogels. Proc. Natl Acad. Sci. USA 112, E3757 (2015).
Yofe, A. D. Physics at surfaces. Contemp. Phys. 29, 411–414 (1988).
Staunton, J. R., Doss, B. L., Lindsay, S. & Ros, R. Correlating confocal microscopy and atomic force indentation reveals metastatic cancer cells stiffen during invasion into collagen I matrices. Sci. Rep. 6, 19686 (2016).
Rheinlaender, J. et al. Cortical cell stiffness is independent of substrate mechanics. Nat. Mater. 19, 1019–1025 (2020).
Symon, K. R. Mechanics. (Addison-Wesley, 1971).
Loebel, C. et al. Metabolic labeling to probe the spatiotemporal accumulation of matrix at the chondrocyte-hydrogel interface. Adv. Funct. Mater. 30, 1909802 (2020).
Dimitriadis, E. K., Horkay, F., Maresca, J., Kachar, B. & Chadwick, R. S. Determination of elastic moduli of thin layers of soft material using the atomic force microscope. Biophys. J. 82, 2798–2810 (2002).
Selby, A., Maldonado-Codina, C. & Derby, B. Influence of specimen thickness on the nanoindentation of hydrogels: measuring the mechanical properties of soft contact lenses. J. Mech. Behav. Biomed. Mater. 35, 144–156 (2014).
Jung, Y. G., Lawn, B. R., Martyniuk, M., Huang, H. & Hu, X. Z. Evaluation of elastic modulus and hardness of thin films by nanoindentation. J. Mater. Res. 19, 3076–3080 (2004).
Sirghi, L., Ponti, J., Broggi, F. & Rossi, F. Probing elasticity and adhesion of live cells by atomic force microscopy indentation. Eur. Biophys. J. 37, 935–945 (2008).
Buxboim, A., Rajagopal, K., Brown, A. E. & Discher, D. E. How deeply cells feel: methods for thin gels. J. Phys. Condens. Matter. 22, 194116 (2010).
Tusan, C. G. et al. Collective cell behavior in mechanosensing of substrate thickness. Biophys. J. 114, 2743–2755 (2018).
Lin, D. C. & Horkay, F. Nanomechanics of polymer gels and biological tissues: a critical review of analytical approaches in the Hertzian regime and beyond. Soft Matter 4, 669–682 (2008).
Carrillo, F. et al. Nanoindentation of polydimethylsiloxane elastomers: Effect of crosslinking, work of adhesion, and fluid environment on elastic modulus. J. Mater. Res. 20, 2820–2830 (2005).
Garcia, P. D., Guerrero, C. R. & Garcia, R. Nanorheology of living cells measured by AFM-based force-distance curves. Nanoscale 12, 9133–9143 (2020).
Efremov, Y. M., Okajima, T. & Raman, A. Measuring viscoelasticity of soft biological samples using atomic force microscopy. Soft Matter 16, 64–81 (2020).
Gautier, H. O. B. et al. in Methods in Cell Biology Vol. 125 211–235 (Academic Press, 2015).
Flory, P. J. Principles of Polymer Chemistry (Cornell University Press, 1953).
Offeddu, G. S., Axpe, E., Harley, B. A. C. & Oyen, M. L. Relationship between permeability and diffusivity in polyethylene glycol hydrogels. AIP Adv 8, 105006 (2018).
Oyen, M. L. Nanoindentation of hydrated materials and tissues. Curr. Opin. Solid State Mater. Sci. 19, 317–323 (2015).
Sader, J. E., Chon, J. W. M. & Mulvaney, P. Calibration of rectangular atomic force microscope cantilevers. Rev. Sci. Instrum. 70, 3967–3969 (1999).
Cleveland, J. P., Manne, S., Bocek, D. & Hansma, P. K. A nondestructive method for determining the spring constant of cantilevers for scanning force microscopy. Rev. Sci. Instrum. 64, 403–405 (1993).
Torii, A., Sasaki, M., Hane, K. & Okuma, S. A method for determining the spring constant of cantilevers for atomic force microscopy. Meas. Sci. Technol. 7, 179–184 (1996).
Gibson, C. T., Watson, G. S. & Myhra, S. Determination of the spring constants of probes for force microscopy/spectroscopy. Nanotechnology 7, 259–262 (1996).
Gates, R. S. & Reitsma, M. G. Precise atomic force microscope cantilever spring constant calibration using a reference cantilever array. Rev. Sci. Instrum. 78, 086101 (2007).
Hutter, J. L. & Bechhoefer, J. Calibration of atomic-force microscope tips. Rev. Sci. Instrum 64, 1868–1873 (1993).
Palacio, M. L. B. & Bhushan, B. Normal and lateral force calibration techniques for AFM cantilevers. Crit. Rev. Solid State Mater. Sci. 35, 73–104 (2010).
Schillers, H. et al. Standardized nanomechanical atomic force microscopy procedure (SNAP) for measuring soft and biological samples. Sci. Rep. 7, 5117 (2017).
Oyen, M. L. & Cook, R. F. A practical guide for analysis of nanoindentation data. J. Mech. Behav. Biomed. Mater. 2, 396–407 (2009).
Oliver, W. C. & Pharr, G. M. An improved technique for determining hardness and elastic modulus using load and displacement sensing indentation experiments. J. Mater. Res. 7, 1564–1583 (1992).
Kohn, J. C. & Ebenstein, D. M. Eliminating adhesion errors in nanoindentation of compliant polymers and hydrogels. J. Mech. Behav. Biomed. Mater. 20, 316–326 (2013).
Li, M., Liu, L., Xi, N. & Wang, Y. Nanoscale monitoring of drug actions on cell membrane using atomic force microscopy. Acta Pharmacol. Sin. 36, 769–782 (2015).
McCracken, K. W., Howell, J. C., Wells, J. M. & Spence, J. R. Generating human intestinal tissue from pluripotent stem cells in vitro. Nat. Protoc. 6, 1920–1928 (2014).
Tse, J. R. & Engler, A. J. Preparation of hydrogel substrates with tunable mechanical properties. Curr. Protoc. Stem Cell Biol. 47, 10.16.11–10.16.16 (2010).
Shu, X. Z., Liu, Y., Luo, Y., Roberts, M. C. & Prestwich, G. D. Disulfide cross-linked hyaluronan hydrogels. Biomacromolecules 3, 1304–1311 (2002).
Ferreira, S. A. et al. Neighboring cells override 3D hydrogel matrix cues to drive human MSC quiescence. Biomaterials 176, 13–23 (2018).
Kloxin, A. M., Kasko, A. M., Salinas, C. N. & Anseth, K. S. Photodegradable hydrogels for dynamic tuning of physical and chemical properties. Science 324, 59 (2009).
Lutolf, M. P. et al. Synthetic matrix metalloproteinase-sensitive hydrogels for the conduction of tissue regeneration: engineering cell-invasion characteristics. Proc. Natl Acad. Sci. USA 100, 5413 (2003).
Burnham, N. A. et al. Comparison of calibration methods for atomic-force microscopy cantilevers. Nanotechnology 14, 1–6 (2003).
Kain, L. et al. Calibration of colloidal probes with atomic force microscopy for micromechanical assessment. J. Mech. Behav. Biomed. Mater. 85, 225–236 (2018).
Chighizola, M., Puricelli, L., Bellon, L. & Podestà, A. Large colloidal probes for atomic force microscopy: fabrication and calibration issues. J. Mol. Recognit. 34, e2879 (2020).
Butt, H. J. & Jaschke, M. Calculation of thermal noise in atomic force microscopy. Nanotechnology 6, 1–7 (1995).
Acknowledgements
M.D.A.N. acknowledges funding from the London Interdisciplinary Doctoral Programme, which is funded by the BBSRC. S.A.F. acknowledges a Springboard Fellowship from the Imperial College London Institutional Strategic Support Fund, which was established with funding from the Wellcome Trust. G.M.J. acknowledges a PhD studentship from the Wellcome Trust (203757/Z/16/A). L.B. acknowledges funding from a University College London Impact Award and industrial sponsorships. E.G. acknowledges a Philip Leverhulme Prize from the Leverhulme Trust. This work was partly funded by generous support from the Rosetrees Trust. The authors are especially grateful for technical support from R. Thorogate at London Centre for Nanotechnology and to T. Ahmed and S. T. Lust for helpful conversations regarding the MATLAB code.
Author information
Authors and Affiliations
Contributions
M.D.A.N., S.A.F. and G.M.J. developed experimental protocols, designed and conducted experiments, and analyzed the data. M.D.A.N., S.A.F., G.M.J., L.B. and E.G. conceived the ideas and contributed to experimental interpretation. All authors wrote and revised the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Protocols thanks the anonymous reviewers for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key references using this protocol
Ferreira, S. A. et al. Nat. Commun. 9, 4049 (2018): https://doi.org/10.1038/s41467-018-06183-4
Foyt, D. A. et al. Acta Biomater. 89, 73–83 (2019): https://doi.org/10.1016/j.actbio.2019.03.002
Jowett, G. M. et al. Nat. Mater. 20, 250–259 (2021): https://doi.org/10.1038/s41563-020-0783-8
Supplementary information
Supplementary Note 1
Supplementary Note and instructions for executing the MATLAB code.
Supplementary Software 1
MATLAB code for identifying the contact point of F-D curves
Supplementary Software 2
MATLAB code for calculating E using the Oliver-Pharr model
Rights and permissions
About this article
Cite this article
Norman, M.D.A., Ferreira, S.A., Jowett, G.M. et al. Measuring the elastic modulus of soft culture surfaces and three-dimensional hydrogels using atomic force microscopy. Nat Protoc 16, 2418–2449 (2021). https://doi.org/10.1038/s41596-021-00495-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-021-00495-4
This article is cited by
-
Profiling native pulmonary basement membrane stiffness using atomic force microscopy
Nature Protocols (2024)
-
Cardiac fibroblasts and mechanosensation in heart development, health and disease
Nature Reviews Cardiology (2023)
-
The extracellular matrix mechanics in the vasculature
Nature Cardiovascular Research (2023)
-
Leaf-venation-directed cellular alignment for macroscale cardiac constructs with tissue-like functionalities
Nature Communications (2023)
-
HG-Induced sEVs Mediate Biomechanics of HK-2 Cells
Nanomanufacturing and Metrology (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.