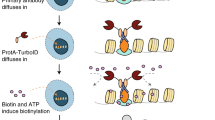
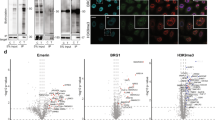
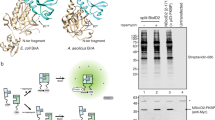
Abstract
This protocol describes the use of TurboID and split-TurboID in proximity labeling applications for mapping protein–protein interactions and subcellular proteomes in live mammalian cells. TurboID is an engineered biotin ligase that uses ATP to convert biotin into biotin–AMP, a reactive intermediate that covalently labels proximal proteins. Optimized using directed evolution, TurboID has substantially higher activity than previously described biotin ligase–related proximity labeling methods, such as BioID, enabling higher temporal resolution and broader application in vivo. Split-TurboID consists of two inactive fragments of TurboID that can be reconstituted through protein–protein interactions or organelle–organelle interactions, which can facilitate greater targeting specificity than full-length enzymes alone. Proteins biotinylated by TurboID or split-TurboID are then enriched with streptavidin beads and identified by mass spectrometry. Here, we describe fusion construct design and characterization (variable timing), proteomic sample preparation (5–7 d), mass spectrometric data acquisition (2 d), and proteomic data analysis (1 week).
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Huber, L. A., Pfaller, K. & Vietor, I. Organelle proteomics: implications for subcellular fractionation in proteomics. Circ. Res. 92, 962–968 (2003).
Puig, O. et al. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods 24, 218–229 (2001).
Stasyk, T. & Huber, L. A. Zooming in: fractionation strategies in proteomics. Proteomics 4, 3704–3716 (2004).
Lee, W. C. & Lee, K. H. Applications of affinity chromatography in proteomics. Anal. Biochem. 324, 1–10 (2004).
Gingras, A. C., Abe, K. T. & Raught, B. Getting to know the neighborhood: using proximity-dependent biotinylation to characterize protein complexes and map organelles. Curr. Opin. Chem. Biol. 48, 44–54 (2019).
Branon, T. C. et al. Efficient proximity labeling in living cells and organisms with TurboID. Nat. Biotechnol. 36, 880–887 (2018).
Cho, K. F. et al. Split-TurboID enables contact-dependent proximity labeling in cells. Proc. Natl Acad. Sci. USA 117, 12143–12154 (2020).
Udeshi, N. D. et al. Antibodies to biotin enable large-scale detection of biotinylation sites on proteins. Nat. Methods 14, 1167–1170 (2017).
Fazal, F. M. et al. Atlas of subcellular RNA localization revealed by APEX-Seq. Cell 178, 473–490.e26 (2019).
Myers, S. A. et al. Discovery of proteins associated with a predefined genomic locus via dCas9-APEX-mediated proximity labeling. Nat. Methods 15, 437–439 (2018).
Michalski, A. et al. Mass spectrometry-based proteomics using Q Exactive, a high-performance benchtop quadrupole Orbitrap mass spectrometer. Mol. Cell. Proteom. 10, M111.011015 (2011).
Eliuk, S. & Makarov, A. Evolution of Orbitrap mass spectrometry instrumentation. Annu. Rev. Anal. Chem. 8, 61–80 (2015).
Tyanova, S., Temu, T. & Cox, J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 11, 2301–2319 (2016).
Lam, S. S. et al. Directed evolution of APEX2 for electron microscopy and proximity labeling. Nat. Methods 12, 51–54 (2014).
Rhee, H. W. et al. Proteomic mapping of mitochondria in living cells via spatially restricted enzymatic tagging. Science 339, 1328–1331 (2013).
Mortensen, A. & Skibsted, L. H. Importance of carotenoid structure in radical-scavenging reactions. J. Agric. Food Chem. 45, 2970–2977 (1997).
Wishart, J. F. & Rao, B. S. M. Recent Trends in Radiation Chemistry (World Scientific, 2010). https://doi.org/10.1142/7413
Martell, J. D. et al. Engineered ascorbate peroxidase as a genetically encoded reporter for electron microscopy. Nat. Biotechnol. 30, 1143–1148 (2012).
Rodríguez-López, J. N. et al. Mechanism of reaction of hydrogen peroxide with horseradish peroxidase: identification of intermediates in the catalytic cycle. J. Am. Chem. Soc. 123, 11838–11847 (2001).
Loh, K. H. et al. Proteomic analysis of unbounded cellular compartments: synaptic clefts. Cell 166, 1295–1307.e21 (2016).
Bar, D. Z. et al. Biotinylation by antibody recognition—a method for proximity labeling. Nat. Methods 15, 127–133 (2018).
Honke, K. & Kotani, N. The enzyme-mediated activation of radical source reaction: a new approach to identify partners of a given molecule in membrane microdomains. J. Neurochem 116, 690–695 (2011).
Li, X.-W. et al. New insights into the DT40 B cell receptor cluster using a proteomic proximity labeling assay. J. Biol. Chem. 289, 14434–14447 (2014).
Paek, J. et al. Multidimensional tracking of GPCR signaling via peroxidase-catalyzed proximity labeling. Cell 169, 338–349.e11 (2017).
Lobingier, B. T. et al. An approach to spatiotemporally resolve protein interaction networks in living cells. Cell 169, 350–360.e12 (2017).
Roux, K. J., Kim, D. I., Raida, M. & Burke, B. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J. Cell Biol. 196, 801–810 (2012).
Kim, D. I. et al. An improved smaller biotin ligase for BioID proximity labeling. Mol. Biol. Cell 27, 1188–1196 (2016).
Ramanathan, M. et al. RNA-protein interaction detection in living cells. Nat. Methods 15, 207–212 (2018).
Choi-Rhee, E., Schulman, H. & Cronan, J. E. Promiscuous protein biotinylation by Escherichia coli biotin protein ligase. Protein Sci. 13, 3043–3050 (2008).
Kim, D. I. et al. Probing nuclear pore complex architecture with proximity-dependent biotinylation. Proc. Natl Acad. Sci. USA 111, E2453–E2461 (2014).
Kido, K. et al. Airid, a novel proximity biotinylation enzyme, for analysis of protein–protein interactions. eLife 9, e54983 (2020).
Birendra, K. C. et al. VRK2A is an A-type lamin-dependent nuclear envelope kinase that phosphorylates BAF. Mol. Biol. Cell 28, 2241–2250 (2017).
Redwine, W. B. et al. The human cytoplasmic dynein interactome reveals novel activators of motility. eLife 6, e28257 (2017).
Jung, E. M. et al. Arid1b haploinsufficiency disrupts cortical interneuron development and mouse behavior. Nat. Neurosci. 20, 1694–1707 (2017).
Mair, A., Xu, S. L., Branon, T. C., Ting, A. Y. & Bergmann, D. C. Proximity labeling of protein complexes and cell type specific organellar proteomes in Arabidopsis enabled by TurboID. eLife 8, e47864 (2019).
Zhang, Y. et al. TurboID-based proximity labeling reveals that UBR7 is a regulator of N NLR immune receptor-mediated immunity. Nat. Commun. 10, 3252 (2019).
Larochelle, M., Bergeron, D., Arcand, B. & Bachand, F. Proximity-dependent biotinylation mediated by TurboID to identify protein-protein interaction networks in yeast. J. Cell Sci. 132, jcs232249 (2019).
Struk, S. et al. Exploring the protein–protein interaction landscape in plants. Plant Cell Environ. 42, 387–409 (2019).
Opitz, N. et al. Capturing the Asc1p/receptor for activated C kinase 1 (RACK1) microenvironment at the head region of the 40s ribosome with quantitative BioID in yeast. Mol. Cell. Proteom. 16, 2199–2218 (2017).
Uezu, A. et al. Identification of an elaborate complex mediating postsynaptic inhibition. Science 353, 1123–1129 (2016).
Lin, Q. et al. Screening of proximal and interacting proteins in rice protoplasts by proximity-dependent biotinylation. Front. Plant Sci. 8, 749 (2017).
Khan, M., Youn, J. Y., Gingras, A. C., Subramaniam, R. & Desveaux, D. In planta proximity dependent biotin identification (BioID). Sci. Rep. 8, 1123 (2018).
Conlan, B., Stoll, T., Gorman, J. J., Saur, I. & Rathjen, J. P. Development of a rapid in planta bioid system as a probe for plasma membrane-associated immunity proteins. Front. Plant Sci. 9, 1882 (2018).
Roux, K. J., Kim, D. I., Burke, B. & May, D. G. BioID: a screen for protein-protein interactions. Curr. Protoc. Protein Sci. 91, 19.23.1–19.23.15 (2018).
May, D. G., Scott, K. L., Campos, A. R. & Roux, K. J. Comparative application of BioID and TurboID for protein-proximity biotinylation. Cells 9, 1070 (2020).
Chapman-Smith, A. & Cronan, J. E. Jr Molecular biology of biotin attachment to proteins. J. Nutr. 129, 477S–484S (1999).
Han, Y. et al. Directed evolution of split APEX2 peroxidase. ACS Chem. Biol. 14, 619–635 (2019).
Martell, J. D. et al. A split horseradish peroxidase for the detection of intercellular protein-protein interactions and sensitive visualization of synapses. Nat. Biotechnol. 34, 774–780 (2016).
De Munter, S. et al. Split-BioID: a proximity biotinylation assay for dimerization-dependent protein interactions. FEBS Lett. 591, 415–424 (2017).
Schopp, I. M. et al. Split-BioID a conditional proteomics approach to monitor the composition of spatiotemporally defined protein complexes. Nat. Commun. 8, 15690 (2017).
Kwak, C. et al. Contact-ID, a new tool for profiling organelle contact site, reveals proteins of mitochondrial-associated membrane formation. Proc. Natl Acad. Sci. USA 117, 12109–12120 (2020).
McClellan, D. et al. Growth factor independence 1B-mediated transcriptional repression and lineage allocation require lysine-specific demethylase 1-dependent recruitment of the BHC complex. Mol. Cell. Biol. 39, e00020-19 (2019).
Lambert, J. P. et al. Interactome rewiring following pharmacological targeting of BET bromodomains. Mol. Cell 73, 621–638.e17 (2019).
Dingar, D. et al. BioID identifies novel c-MYC interacting partners in cultured cells and xenograft tumors. J. Proteom. 118, 95–111 (2015).
Couzens, A. L. et al. Protein interaction network of the mammalian hippo pathway reveals mechanisms of kinase-phosphatase interactions. Sci. Signal. 6, rs15–rs15 (2013).
Gupta, G. D. et al. A dynamic protein interaction landscape of the human centrosome-cilium interface. Cell 163, 1484–1499 (2015).
Youn, J. Y. et al. High-density proximity mapping reveals the subcellular organization of mRNA-associated granules and bodies. Mol. Cell 69, 517–532.e11 (2018).
Firat-Karalar, E. N., Rauniyar, N., Yates, J. R. & Stearns, T. Proximity interactions among centrosome components identify regulators of centriole duplication. Curr. Biol. 24, 664–670 (2014).
Chou, C. C. et al. TDP-43 pathology disrupts nuclear pore complexes and nucleocytoplasmic transport in ALS/FTD. Nat. Neurosci. 21, 228–239 (2018).
Kabeiseman, E. J., Cichos, K. H. & Moore, E. R. The eukaryotic signal sequence, YGRL, targets the chlamydial inclusion. Front. Cell. Infect. Microbiol. 4, 129 (2014).
Mojica, S. A. et al. SINC, a type III secreted protein of Chlamydia psittaci, targets the inner nuclear membrane of infected cells and uninfected neighbors. Mol. Biol. Cell 26, 1918–1934 (2015).
Le Sage, V., Cinti, A., Valiente-Echeverría, F. & Mouland, A. J. Proteomic analysis of HIV-1 Gag interacting partners using proximity-dependent biotinylation. Virol. J. 12, 138 (2015).
Ritchie, C., Cylinder, I., Platt, E. J. & Barklis, E. Analysis of HIV-1 Gag protein interactions via biotin ligase tagging. J. Virol. 89, 3988–4001 (2015).
Kueck, T. et al. Serine phosphorylation of HIV-1 Vpu and its binding to tetherin regulates interaction with clathrin adaptors. PLoS Pathog. 11, e1005141 (2015).
Holthusen, K., Talaty, P. & Everly, D. N. Regulation of latent membrane protein 1 signaling through interaction with cytoskeletal proteins. J. Virol. 89, 7277–7290 (2015).
Coyaud, E. et al. Global interactomics uncovers extensive organellar targeting by Zika virus. Mol. Cell. Proteom. 17, 2242–2255 (2018).
Rider, M. A. et al. The interactome of EBV LMP1 evaluated by proximity-based BioID approach. Virology 516, 55–70 (2018).
Cheerathodi, M. R. & Meckes, D. G. BioID combined with mass spectrometry to study herpesvirus protein–protein interaction networks. Methods Mol. Biol. 2060, 327–341 (2020).
Bradley, P. J., Rayatpisheh, S., Wohlschlegel, J. A. & Nadipuram, S. M. Using BioID for the identification of interacting and proximal proteins in subcellular compartments in Toxoplasma gondii. Methods Mol. Biol. 2071, 323–346 (2020).
Gillingham, A. K., Bertram, J., Begum, F. & Munro, S. In vivo identification of GTPase interactors by mitochondrial relocalization and proximity biotinylation. eLife 8, e45916 (2019).
Hoyer, M. J. et al. A novel class of ER membrane proteins regulates ER-associated endosome fission. Cell 175, 254–265.e14 (2018).
van Vliet, A. R. et al. The ER stress sensor PERK coordinates ER-plasma membrane contact site formation through interaction with filamin-A and F-actin remodeling. Mol. Cell 65, 885–899.e6 (2017).
Spence, E. F. et al. In vivo proximity proteomics of nascent synapses reveals a novel regulator of cytoskeleton-mediated synaptic maturation. Nat. Commun. 10, 386 (2019).
Feng, W. et al. Identifying the cardiac dyad proteome in vivo by a BioID2 knock-in strategy. Circulation 141, 940–942 (2020).
Hung, V. et al. Spatially resolved proteomic mapping in living cells with the engineered peroxidase APEX2. Nat. Protoc. 11, 456–475 (2016).
Hung, V. et al. Proteomic mapping of the human mitochondrial intermembrane space in live cells via ratiometric APEX tagging. Mol. Cell 55, 332–341 (2014).
Han, S. et al. Proximity biotinylation as a method for mapping proteins associated with mtDNA in living cells. Cell Chem. Biol. 24, 404–414 (2017).
Hung, V. et al. Proteomic mapping of cytosol-facing outer mitochondrial and ER membranes in living human cells by proximity biotinylation. eLife 6, e24463 (2017).
Mertins, P. et al. Reproducible workflow for multiplexed deep-scale proteome and phosphoproteome analysis of tumor tissues by liquid chromatography-mass spectrometry. Nat. Protoc. 13, 1632–1661 (2018).
Li, J. et al. Cell-surface proteomic profiling in the fly brain uncovers wiring regulators. Cell 180, 373–386.e15 (2020).
Vandemoortele, G. et al. A well-controlled BioID design for endogenous bait proteins. J. Proteome Res. 18, 95–106 (2019).
Bian, Y. et al. Robust, reproducible and quantitative analysis of thousands of proteomes by micro-flow LC–MS/MS. Nat. Commun. 11, 157 (2020).
Käll, L., Krogh, A. & Sonnhammer, E. L. L. A combined transmembrane topology and signal peptide prediction method. J. Mol. Biol. 338, 1027–1036 (2004).
Ashburner, M. et al. Gene ontology: tool for the unification of biology. Nat. Genet. 25, 25–29 (2000).
Gene Ontology Consortium. Gene Ontology Consortium: going forward. Nucleic Acids Res. 43, D1049–D1056 (2015).
Rappsilber, J., Mann, M. & Ishihama, Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2, 1896–1906 (2007).
Bateman, A. et al. UniProt: the universal protein knowledgebase. Nucleic Acids Res 45, D158–D169 (2017).
Lee, S. Y. et al. APEX fingerprinting reveals the subcellular localization of proteins of interest. Cell Rep. 15, 1837–1847 (2016).
Cho, I. T. et al. Ascorbate peroxidase proximity labeling coupled with biochemical fractionation identifies promoters of endoplasmic reticulum–mitochondrial contacts. J. Biol. Chem. 292, 16382–16392 (2017).
Cao, Q. et al. PAQR3 regulates endoplasmic reticulum-to-Golgi trafficking of COPII vesicle via interaction with Sec13/Sec31 coat proteins. iScience 9, 382–398 (2018).
Le Guerroué, F. et al. Autophagosomal content profiling reveals an LC3C-dependent piecemeal mitophagy pathway. Mol. Cell 68, 786–796.e6 (2017).
Mick, D. U. et al. Proteomics of primary cilia by proximity labeling. Dev. Cell 35, 497–512 (2015).
Santin, Y. G. et al. In vivo TssA proximity labelling during type VI secretion biogenesis reveals TagA as a protein that stops and holds the sheath. Nat. Microbiol. 3, 1304–1313 (2018).
Mannix, K. M., Starble, R. M., Kaufman, R. S. & Cooley, L. Proximity labeling reveals novel interactomes in live Drosophila tissue. Development 146, dev176644 (2019).
Liu, G. et al. Mechanism of adrenergic CaV1.2 stimulation revealed by proximity proteomics. Nature 577, 695–700 (2020).
Chojnowski, A. et al. Progerin reduces LAP2α-telomere association in Hutchinson-Gilford progeria. eLife 4, 1–21 (2015).
Cross, S. H. et al. The nanophthalmos protein TMEM98 inhibits MYRF self-cleavage and is required for eye size specification. PLoS Genet 16, e1008583 (2020).
Pagac, M. et al. SEIPIN regulates lipid droplet expansion and adipocyte development by modulating the activity of glycerol-3-phosphate acyltransferase. Cell Rep. 17, 1546–1559 (2016).
Cole, A. et al. Inhibition of the mitochondrial protease ClpP as a therapeutic strategy for human acute myeloid leukemia. Cancer Cell 27, 864–876 (2015).
Janer, A. et al. SLC 25A46 is required for mitochondrial lipid homeostasis and cristae maintenance and is responsible for Leigh syndrome. EMBO Mol. Med. 8, 1019–1038 (2016).
Antonicka, H. et al. A pseudouridine synthase module is essential for mitochondrial protein synthesis and cell viability. EMBO Rep. 18, 28–38 (2017).
Van Itallie, C. M. et al. Biotin ligase tagging identifies proteins proximal to E-cadherin, including lipoma preferred partner, a regulator of epithelial cell-cell and cell-substrate adhesion. J. Cell Sci. 127, 885–895 (2014).
Guo, Z. et al. E-cadherin interactome complexity and robustness resolved by quantitative proteomics. Sci. Signal. 7, rs7 (2014).
Hua, R. et al. VAPs and ACBD5 tether peroxisomes to the ER for peroxisome maintenance and lipid homeostasis. J. Cell Biol. 216, 367–377 (2017).
Chan, C. J. et al. BioID performed on Golgi enriched fractions identify C10orf76 as a GBF1 binding protein essential for Golgi maintenance and secretion. Mol. Cell. Proteom. 18, 2285–2297 (2019).
Opitz, N. et al. Capturing the Asc1p/ R eceptor for A ctivated C K inase 1 (RACK1) microenvironment at the head region of the 40S ribosome with quantitative BioID in yeast. Mol. Cell. Proteom. 16, 2199–2218 (2017).
Domsch, K. et al. The hox transcription factor ubx stabilizes lineage commitment by suppressing cellular plasticity in Drosophila. eLife 8, e42675 (2019).
Bagchi, P., Torres, M., Qi, L. & Tsai, B. Selective EMC subunits act as molecular tethers of intracellular organelles exploited during viral entry. Nat. Commun. 11, 1127 (2020).
Yoshinaka, T. et al. Structural basis of mitochondrial scaffolds by prohibitin complexes: insight into a role of the coiled-coil region. iScience 19, 1065–1078 (2019).
Callegari, S. et al. A MICOS–TIM22 association promotes carrier import into human mitochondria. J. Mol. Biol. 431, 2835–2851 (2019).
Chen, Z. et al. Global phosphoproteomic analysis reveals ARMC10 as an AMPK substrate that regulates mitochondrial dynamics. Nat. Commun. 10, 104 (2019).
Liu, L., Doray, B. & Kornfeld, S. Recycling of Golgi glycosyltransferases requires direct binding to coatomer. Proc. Natl Acad. Sci. USA. 115, 8984–8989 (2018).
Mirza, A. N. et al. LAP2 proteins chaperone GLI1 movement between the lamina and chromatin to regulate transcription. Cell 176, 198–212.e15 (2019).
Acknowledgements
This work was supported by NIH R01-DK121409 (to A.Y.T. and S.A.C.) and the Stanford Wu Tsai Neurosciences Institute Big Ideas Initiative (to A.Y.T.). K.F.C. was supported by NIH Training Grant 2T32CA009302-41 and the Blavatnik Graduate Fellowship. T.C.B. is a Robert Black Fellow of the Damon Runyon Cancer Research Foundation (DRG-2391-20). A.Y.T. is an investigator of the Chan Zuckerberg Biohub.
Author information
Authors and Affiliations
Contributions
K.F.C., T.C.B, N.D.U., S.A.M., S.A.C., and A.Y.T. contributed to the writing and editing of the manuscript.
Corresponding author
Ethics declarations
Competing interests
A.Y.T. and T.C.B. have filed a patent application covering some aspects of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key references using this protocol:
Branon, T. C. et al. Nat. Biotechnol. 36, 880–887 (2018): https://www.nature.com/articles/nbt.4201
Cho, K. F. et al. Proc. Natl Acad. Sci. USA 117, 12143–12154 (2020): https://www.pnas.org/content/117/22/12143
Supplementary information
Supplementary Table 1
Human proteome of proteins, annotated by whether each protein was previously detected in a PL proteomic experiment from our lab (regions include: mitochondrial matrix6,15, mitochondrial intermembrane space76, mitochondrial nucleoid77, ER membrane6,7,78, outer mitochondrial membrane7,78, ER-mitochondria contact sites7,78, nucleus6, synaptic cleft20, and cytosol6,7,78). For each protein, the compartment(s) in which they were detected are listed.
Supplementary Table 2
Compilation of data from previous PL proteomic mapping experiments performed by our lab, categorized by organelle/region of interest (each tab is a different subcellular compartment). In each tab, the relevant studies and corresponding enrichment ratios (SILAC, TMT, or iTRAQ) for proteins detected above the respective cutoffs are provided. Data are included for the mitochondrial matrix6,15 (Tab 1), mitochondrial intermembrane space76 (Tab 2), mitochondrial nucleoid77 (Tab 3), ER membrane6,7,78 (Tab 4), outer mitochondrial membrane7,78 (Tab 5), ER-mitochondria contact sites7,78 (Tab 6), nucleus6 (Tab 7), synaptic cleft20 (Tab 8), and cytosol6,7,78 (Tab 9).
Rights and permissions
About this article
Cite this article
Cho, K.F., Branon, T.C., Udeshi, N.D. et al. Proximity labeling in mammalian cells with TurboID and split-TurboID. Nat Protoc 15, 3971–3999 (2020). https://doi.org/10.1038/s41596-020-0399-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-020-0399-0
This article is cited by
-
Overcoming genetic and cellular complexity to study the pathophysiology of X-linked intellectual disabilities
Journal of Neurodevelopmental Disorders (2024)
-
Canalizing cell fate by transcriptional repression
Molecular Systems Biology (2024)
-
In vivo identification of Drosophila rhodopsin interaction partners by biotin proximity labeling
Scientific Reports (2024)
-
The diversification of methods for studying cell–cell interactions and communication
Nature Reviews Genetics (2024)
-
In vivo identification of astrocyte and neuron subproteomes by proximity-dependent biotinylation
Nature Protocols (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.