Abstract
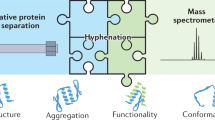
The higher-order structure (HOS) of proteins plays a critical role in their function; therefore, it is important to our understanding of their function that we have as much information as possible about their three-dimensional structure and how it changes with time. Mass spectrometry (MS) has become an important tool for determining protein HOS owing to its high throughput, mid-to-high spatial resolution, low sample amount requirement and broad compatibility with various protein systems. Modern MS-based protein HOS analysis relies, in part, on footprinting, where a reagent reacts ‘to mark’ the solvent-accessible surface of the protein, and MS-enabled proteomic analysis locates the modifications to afford a footprint. Fast photochemical oxidation of proteins (FPOP), first introduced in 2005, has become a powerful approach for protein footprinting. Laser-induced hydrogen peroxide photolysis generates hydroxyl radicals that react with solvent-accessible side chains (14 out of 20 amino acid side chains) to fulfill the footprinting. The reaction takes place at sub-milliseconds, faster than most of labeling-induced protein conformational changes, thus enabling a ‘snapshot’ of protein HOS in solution. As a result, FPOP has been employed in solving several important problems, including mapping epitopes, following protein aggregation, locating small molecule binding, measuring ligand-binding affinity, monitoring protein folding and unfolding and determining hidden conformational changes invisible to other methods. Broader adoption will be promoted by dissemination of the technical details for assembling the FPOP platform and for dealing with the complexities of analyzing FPOP data. In this protocol, we describe the FPOP platform, the conditions for successful footprinting and its examination by mass measurements of the intact protein, the post-labeling sample handling and digestion, the liquid chromatography–tandem MS analysis of the digested sample and the data analysis with Protein Metrics Suite. This protocol is intended not only as a guide for investigators trying to establish an FPOP platform in their own lab but also for those willing to incorporate FPOP as an additional tool in addressing their questions of interest.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
A calmodulin data set that contains a global-level measurement by Bruker maXis and a peptide- and residue-level measurement by Thermo Fisher Scientific Q Exactive is freely available online in the Mendeley data (https://data.mendeley.com/) with DOI: 10.17632/xfd5sh76pm.1.
Software availability
The Protein Metrics Suite is commercially available at https://www.proteinmetrics.com.
References
Anfinsen, C. B., Haber, E., Sela, M. & White, F. H. The kinetics of formation of native ribonuclease during oxidation of the reduced polypeptide chain. Proc. Natl Acad. Sci. USA 47, 1309–1314 (1961).
Drenth, J. Principles of Protein X-Ray Crystallography (Springer Science & Business Media, 2007).
Wüthrich, K. The way to NMR structures of proteins. Nat. Struct. Biol. 8, 923–925 (2001).
McPherson, A. Introduction to protein crystallization. Methods 34, 254–265 (2004).
Cheng, Y. Single-particle cryo-EM at crystallographic resolution. Cell 161, 450–457 (2015).
Bai, X.-c, McMullan, G. & Scheres, S. H. W. How cryo-EM is revolutionizing structural biology. Trends Biochem. Sci. 40, 49–57 (2015).
Merk, A. et al. Breaking cryo-EM resolution barriers to facilitate drug discovery. Cell 165, 1698–1707 (2016).
Greenfield, N. J. Using circular dichroism spectra to estimate protein secondary structure. Nat. Prot. 1, 2876–2890 (2006).
Noble, J. E.& Bailey, M. J. A. in Methods in Enzymology, Vol. 463 (eds Burgess, R. R. & Deutscher, M. P.) Ch. 8 73–95 (Academic Press, 2009).
Kong, J. & Yu, S. Fourier transform infrared spectroscopic analysis of protein secondary structures. Acta Biochim. Biophys. Sin. 39, 549–559 (2007).
Stetefeld, J., McKenna, S. A. & Patel, T. R. Dynamic light scattering: a practical guide and applications in biomedical sciences. Biophys. Rev. 8, 409–427 (2016).
Dančík, V., Addona, T. A., Clauser, K. R., Vath, J. E. & Pevzner, P. A. De novo peptide sequencing via tandem mass spectrometry. J. Comput. Biol. 6, 327–342 (1999).
Witze, E. S., Old, W. M., Resing, K. A. & Ahn, N. G. Mapping protein post-translational modifications with mass spectrometry. Nat. Methods 4, 798–806 (2007).
Liu, X. R., Zhang, M. M. & Gross, M. L. Mass spectrometry-based protein footprinting for higher-order structure analysis: fundamentals and applications. Chem. Rev. 120, 4335–4454 (2020).
Mendoza, V. L. & Vachet, R. W. Probing protein structure by amino acid-specific covalent labeling and mass spectrometry. Mass Spectrom. Rev. 28, 785–815 (2009).
Konermann, L., Pan, J. & Liu, Y.-H. Hydrogen exchange mass spectrometry for studying protein structure and dynamics. Chem. Soc. Rev. 40, 1224–1234 (2011).
Maleknia, S. D. & Downard, K. M. Advances in radical probe mass spectrometry for protein footprinting in chemical biology applications. Chem. Soc. Rev. 43, 3244–3258 (2014).
Bolla, J. R., Agasid, M. T., Mehmood, S. & Robinson, C. V. Membrane protein–lipid interactions probed using mass spectrometry. Annu. Rev. Biochem. 88, 85–111 (2019).
Xu, G. & Chance, M. R. Hydroxyl radical-mediated modification of proteins as probes for structural proteomics. Chem. Rev. 107, 3514–3543 (2007).
Kaur, U. et al. Evolution of structural biology through the lens of mass spectrometry. Anal. Chem. 91, 142–155 (2019).
Hernández, H. & Robinson, C. V. Determining the stoichiometry and interactions of macromolecular assemblies from mass spectrometry. Nat. Prot. 2, 715 (2007).
Benesch, J. L. P., Ruotolo, B. T., Simmons, D. A. & Robinson, C. V. Protein complexes in the gas phase: technology for structural genomics and proteomics. Chem. Rev. 107, 3544–3567 (2007).
Sinz, A. Chemical cross-linking and mass spectrometry to map three-dimensional protein structures and protein–protein interactions. Mass Spectrom. Rev. 25, 663–682 (2006).
Uetrecht, C., Rose, R. J., van Duijn, E., Lorenzen, K. & Heck, A. J. R. Ion mobility mass spectrometry of proteins and protein assemblies. Chem. Soc. Rev. 39, 1633–1655 (2010).
Zhang, M. M. et al. An integrated approach for determining a protein–protein binding interface in solution and an evaluation of hydrogen–deuterium exchange kinetics for adjudicating candidate docking models. Anal. Chem. 91, 15709–15717 (2019).
Zhang, M. M. et al. Epitope and paratope mapping of PD-1/nivolumab by mass spectrometry–based hydrogen–deuterium xchange, cross-linking, and molecular docking. Anal. Chem. 92, 9086–9094 (2020).
Kelleher, N. L. et al. Top down versus bottom up protein characterization by tandem high-resolution mass spectrometry. J. Am. Chem. Soc. 121, 806–812 (1999).
Espino, J. A. & Jones, L. M. Illuminating biological interactions with in vivo protein footprinting. Anal. Chem. 91, 6577–6584 (2019).
West, G. M. et al. Quantitative proteomics approach for identifying protein–drug interactions in complex mixtures using protein stability measurements. Proc. Natl Acad. Sci. USA 107, 9078–9082 (2010).
Jin, L., Wang, D., Gooden, D. M., Ball, C. H. & Fitzgerald, M. C. Targeted mass spectrometry-based approach for protein–ligand binding analyses in complex biological mixtures using a phenacyl bromide modification strategy. Anal. Chem. 88, 10987–10993 (2016).
Katta, V., Chait, B. T. & Carr, S. Conformational changes in proteins probed by hydrogen-exchange electrospray-ionization mass spectrometry. Rapid Commun. Mass Spectrom. 5, 214–217 (1991).
Zhang, Z. & Smith, D. L. Determination of amide hydrogen exchange by mass spectrometry: a new tool for protein structure elucidation. Protein Sci. 2, 522–531 (1993).
Englander, S. W. & Kallenbach, N. R. Hydrogen exchange and structural dynamics of proteins and nucleic acids. Q. Rev. Biophys. 16, 521–655 (1983).
Schanda, P. & Brutscher, B. Very fast two-dimensional NMR spectroscopy for real-time investigation of dynamic events in proteins on the time scale of seconds. J. Am. Chem. Soc. 127, 8014–8015 (2005).
Wales, T. E. & Engen, J. R. Hydrogen exchange mass spectrometry for the analysis of protein dynamics. Mass Spectrom. Rev. 25, 158–170 (2006).
Engen, J. R. Analysis of protein conformation and dynamics by hydrogen/deuterium exchange MS. Anal. Chem. 81, 7870–7875 (2009).
Anderson, K. W., Gallagher, E. S. & Hudgens, J. W. Automated removal of phospholipids from membrane proteins for h/d exchange mass spectrometry workflows. Anal. Chem. 90, 6409–6412 (2018).
Möller, I. R. et al. Improving the sequence coverage of integral membrane proteins during hydrogen/deuterium exchange mass spectrometry experiments. Anal. Chem. 91, 10970–10978 (2019).
Jensen, P. F. et al. Removal of N-linked glycosylations at acidic pH by PNGase A facilitates hydrogen/deuterium exchange mass spectrometry analysis of N-linked glycoproteins. Anal. Chem. 88, 12479–12488 (2016).
Zhang, J.-G., Reid, G. E., Moritz, R. L., Ward, L. D. & Simpson, R. J. Specific covalent modification of the tryptophan residues in murine interleukin-6. Eur. J. Biochem. 217, 55–59 (1993).
Liu, T., Marcinko, T. M., Kiefer, P. A. & Vachet, R. W. Using covalent labeling and mass spectrometry to study protein binding sites of amyloid inhibiting molecules. Anal. Chem. 89, 11583–11591 (2017).
Li, K. S., Shi, L. & Gross, M. L. Mass spectrometry-based fast photochemical oxidation of proteins (FPOP) for higher order structure characterization. Acc. Chem. Res. 51, 736–744 (2018).
Maleknia, S. D., Brenowitz, M. & Chance, M. R. Millisecond radiolytic modification of peptides by synchrotron X-rays identified by mass spectrometry. Anal. Chem. 71, 3965–3973 (1999).
Hambly, D. M. & Gross, M. L. Laser flash photolysis of hydrogen peroxide to oxidize protein solvent-accessible residues on the microsecond timescale. J. Am. Soc. Mass Spectrom. 16, 2057–2063 (2005).
Li, J. et al. Mapping the energetic epitope of an antibody/interleukin-23 interaction with hydrogen/deuterium exchange, fast photochemical oxidation of proteins mass spectrometry, and alanine shave mutagenesis. Anal. Chem. 89, 2250–2258 (2017).
Jones, L. M., B. Sperry, J., A. Carroll, J. & Gross, M. L. Fast photochemical oxidation of proteins for epitope mapping. Anal. Chem. 83, 7657–7661 (2011).
Stocks, B. B. & Konermann, L. Structural characterization of short-lived protein unfolding intermediates by laser-induced oxidative labeling and mass spectrometry. Anal. Chem. 81, 20–27 (2009).
Chen, J., Rempel, D. L. & Gross, M. L. Temperature jump and fast photochemical oxidation probe submillisecond protein folding. J. Am. Chem. Soc. 132, 15502–15504 (2010).
Chen, J., Rempel, D. L., Gau, B. C. & Gross, M. L. Fast photochemical oxidation of proteins and mass spectrometry follow submillisecond protein folding at the amino-acid level. J. Am. Chem. Soc. 134, 18724–18731 (2012).
Li, K. S., Rempel, D. L. & Gross, M. L. Conformational-sensitive fast photochemical oxidation of proteins and mass spectrometry characterize amyloid beta 1–42 aggregation. J. Am. Chem. Soc. 138, 12090–12098 (2016).
Liu, X. R., Zhang, M. M., Rempel, D. L. & Gross, M. L. Protein–ligand interaction by ligand titration, fast photochemical oxidation of proteins and mass spectrometry: LITPOMS. J. Am. Soc. Mass Spectrom. 30, 213–217 (2019).
Liu, X. R., Zhang, M. M., Rempel, D. L. & Gross, M. L. A single approach reveals the composite conformational changes, order of binding, and affinities for calcium binding to calmodulin. Anal. Chem. 91, 5508–5512 (2019).
Liu, X. R., Rempel, D. L. & Gross, M. L. Composite conformational changes of signaling proteins upon ligand binding revealed by a single approach: calcium–calmodulin study. Anal. Chem. 91, 12560–12567 (2019).
Espino, J. A., Mali, V. S. & Jones, L. M. In cell footprinting coupled with mass spectrometry for the structural analysis of proteins in live cells. Anal. Chem. 87, 7971–7978 (2015).
Aprahamian, M. L., Chea, E. E., Jones, L. M. & Lindert, S. Rosetta protein structure prediction from hydroxyl radical protein footprinting mass spectrometry data. Anal. Chem. 90, 7721–7729 (2018).
Aprahamian, M. L. & Lindert, S. Utility of covalent labeling mass spectrometry data in protein structure prediction with Rosetta. J. Chem. Theory Comput. 15, 3410–3424 (2019).
Cheng, M., Zhang, B., Cui, W. & Gross, M. L. Laser-initiated radical trifluoromethylation of peptides and proteins: application to mass-spectrometry-based protein footprinting. Angew. Chem. Int. Ed. 56, 14007–14010 (2017).
Zhang, M. M., Rempel, D. L. & Gross, M. L. A Fast Photochemical Oxidation of Proteins (FPOP) platform for free-radical reactions: the carbonate radical anion with peptides and proteins. Free Radic. Biol. Med. 131, 126–132 (2019).
Zhang, B., Rempel, D. L. & Gross, M. L. Protein footprinting by carbenes on a fast photochemical oxidation of proteins (FPOP) platform. Am. Soc. Mass Spectrom. 27, 552–555 (2016).
Liu, X. R., Zhang, M. M., Zhang, B., Rempel, D. L. & Gross, M. L. Hydroxyl-radical reaction pathways for the fast photochemical oxidation of proteins platform as revealed by 18O isotopic labeling. Anal. Chem. 91, 9238–9245 (2019).
Weisz, D. A., Gross, M. L. & Pakrasi, H. B. Reactive oxygen species leave a damage trail that reveals water channels in Photosystem II. Sci. Adv. 3, eaao3013 (2017).
Heyduk, E. & Heyduk, T. Mapping protein domains involved in macromolecular interactions: a novel protein footprinting approach. Biochemistry 33, 9643–9650 (1994).
Sharp, J. S., Becker, J. M. & Hettich, R. L. Analysis of protein solvent accessible surfaces by photochemical oxidation and mass spectrometry. Anal. Chem. 76, 672–683 (2004).
Aye, T. T., Low, T. Y. & Sze, S. K. Nanosecond laser-induced photochemical oxidation method for protein surface mapping with mass spectrometry. Anal. Chem. 77, 5814–5822 (2005).
Yan, Y. et al. Fast photochemical oxidation of proteins (FPOP) maps the epitope of egfr binding to adnectin. J. Am. Soc. Mass Spectrom. 25, 2084–2092 (2014).
Zhang, H., Gau, B. C., Jones, L. M., Vidavsky, I. & Gross, M. L. Fast photochemical oxidation of proteins for comparing structures of protein–ligand complexes: the calmodulin-peptide model system. Anal. Chem. 83, 311–318 (2011).
Zhang, Y., Rempel, D. L., Zhang, H. & Gross, M. L. An improved fast photochemical oxidation of proteins (FPOP) platform for protein therapeutics. J. Am. Soc. Mass Spectrom. 26, 526–529 (2015).
Garrison, W. M. Reaction mechanisms in the radiolysis of peptides, polypeptides, and proteins. Chem. Rev. 87, 381–398 (1987).
Chen, J., Cui, W., Giblin, D. & Gross, M. L. New protein footprinting: fast photochemical iodination combined with top-down and bottom-up mass spectrometry. J. Am. Soc. Mass Spectrom. 23, 1306–1318 (2012).
Manzi, L. et al. Carbene footprinting accurately maps binding sites in protein–ligand and protein–protein interactions. Nat. Commun. 7, 13288 (2016).
Hambly, D. M. & Gross, M. L. Cold chemical oxidation of proteins. Anal. Chem. 81, 7235–7242 (2009).
Gau, B. C., Sharp, J. S., Rempel, D. L. & Gross, M. L. Fast photochemical oxidation of protein footprints faster than protein unfolding. Anal. Chem. 81, 6563–6571 (2009).
Srikanth, R., Wilson, J. & Vachet, R. W. Correct identification of oxidized histidine residues using electron-transfer dissociation. J. Mass Spectrom. 44, 755–762 (2009).
Li, X., Li, Z., Xie, B. & Sharp, J. S. Improved identification and relative quantification of sites of peptide and protein oxidation for hydroxyl radical footprinting. J. Am. Soc. Mass Spectrom. 24, 1767–1776 (2013).
Acknowledgements
This work was supported by National Institutes of Health National Institute of General Medical Sciences grants 5P41GM103422 and R24GM135766. Instrumentation was supplied by 1S10OD016298-01A1 (to M.L.G.). We are grateful to H. Rohrs for help in identifying parts for the HPLC and to Protein Metrics for software support.
Author information
Authors and Affiliations
Contributions
X.R.L., D.L.R. and M.L.G. developed the protocol. X.R.L. and M.L.G. wrote and edited the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare an ongoing collaboration with Protein Metrics in establishing an HDX data processing platform, a topic that is not related to this protocol. The recommendation of using Protein Metrics as preferred data processing software for FPOP data predates the ongoing collaboration. M.L.G. is an unpaid member of the scientific advisory board of GenNext Technologies, which provides products and services for hydroxyl radical footprinting.
Additional information
Peer review information Nature Protocols thanks Richard W. Vachet and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key references using this protocol
Hambly, D. M. et. al. J. Am. Soc. Mass Spectrom. 16, 2057–2063 (2005): https://doi.org/10.1016/j.jasms.2005.09.008
Espino, J. A. et. al. Anal. Chem. 87, 7971–7978 (2015): https://doi.org/10.1021/acs.analchem.5b01888
Li, K. S. et. al. J. Am Chem. Soc. 138, 12090–12098 (2016): https://doi.org/10.1021/jacs.6b07543
Niu, B. et. al. J. Am. Soc. Mass Spectrom. 28, 389–392 (2017): https://doi.org/10.1007/s13361-016-1552-4
Liu, X. R. et. al. Anal. Chem. 91, 12560–12567 (2019): https://doi.org/10.1021/acs.analchem.9b03491
Supplementary information
Supplementary Information
Supplementary Figs. 1 and 2.
Rights and permissions
About this article
Cite this article
Liu, X.R., Rempel, D.L. & Gross, M.L. Protein higher-order-structure determination by fast photochemical oxidation of proteins and mass spectrometry analysis. Nat Protoc 15, 3942–3970 (2020). https://doi.org/10.1038/s41596-020-0396-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-020-0396-3
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.