Abstract
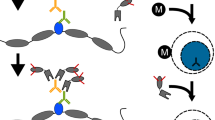
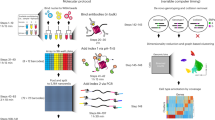
We recently introduced Cleavage Under Targets & Tagmentation (CUT&Tag), an epigenomic profiling strategy in which antibodies are bound to chromatin proteins in situ in permeabilized nuclei. These antibodies are then used to tether the cut-and-paste transposase Tn5. Activation of the transposase simultaneously cleaves DNA and adds adapters (‘tagmentation’) for paired-end DNA sequencing. Here, we introduce a streamlined CUT&Tag protocol that suppresses DNA accessibility artefacts to ensure high-fidelity mapping of the antibody-targeted protein and improves the signal-to-noise ratio over current chromatin profiling methods. Streamlined CUT&Tag can be performed in a single PCR tube, from cells to amplified libraries, providing low-cost genome-wide chromatin maps. By simplifying library preparation CUT&Tag requires less than a day at the bench, from live cells to sequencing-ready barcoded libraries. As a result of low background levels, barcoded and pooled CUT&Tag libraries can be sequenced for as little as $25 per sample. This enables routine genome-wide profiling of chromatin proteins and modifications and requires no special skills or equipment.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Rodriguez-Ubreva, J. & Ballestar, E. Chromatin immunoprecipitation. Methods Mol. Biol. 1094, 309–318 (2014).
Solomon, M. J. & Varshavsky, A. Formaldehyde-mediated DNA-protein crosslinking: a probe for in vivo chromatin structures. Proc. Natl Acad. Sci. USA 82, 6470–6474 (1985).
Rossi, M. J., Lai, W. K. M. & Pugh, B. F. Simplified ChIP-exo assays. Nat. Commun. 9, 2842 (2018).
He, Q., Johnston, J. & Zeitlinger, J. ChIP-nexus enables improved detection of in vivo transcription factor binding footprints. Nat. Biotechnol. 33, 395–401 (2015).
Skene, P. J. & Henikoff, S. A simple method for generating high-resolution maps of genome wide protein binding. eLife 4, e09225 (2015).
Kasinathan, S., Orsi, G. A., Zentner, G. E., Ahmad, K. & Henikoff, S. High-resolution mapping of transcription factor binding sites on native chromatin. Nat. Methods 11, 203–209 (2014).
Ai, S. et al. Profiling chromatin states using single-cell itChIP-seq. Nat. Cell Biol. 21, 1164–1172 (2019).
Grosselin, K. et al. High-throughput single-cell ChIP-seq identifies heterogeneity of chromatin states in breast cancer. Nat. Genet. 51, 1060–1066 (2019).
Schmidl, C., Rendeiro, A. F., Sheffield, N. C. & Bock, C. ChIPmentation: fast, robust, low-input ChIP-seq for histones and transcription factors. Nat. Methods 12, 963–965 (2015).
van Steensel, B. & Henikoff, S. Identification of in vivo DNA targets of chromatin proteins using tethered Dam methyltransferase. Nat. Biotechnol. 18, 424–428 (2000).
Schmid, M., Durussel, T. & Laemmli, U. K. ChIC and ChEC; genomic mapping of chromatin proteins. Mol. Cell 16, 147–157 (2004).
Zentner, G. E., Kasinathan, S., Xin, B., Rohs, R. & Henikoff, S. ChEC-seq kinetics discriminate transcription factor binding sites by DNA sequence and shape in vivo. Nat. Commun. 6, 8733 (2015).
Skene, P. J. & Henikoff, S. An efficient targeted nuclease strategy for high-resolution mapping of DNA binding sites. Elife 6, e21856 (2017).
Janssens, D. H. et al. Automated in situ chromatin profiling efficiently resolves cell types and gene regulatory programs. Epigenetics Chromatin 11, 74 (2018).
Skene, P. J., Henikoff, J. G. & Henikoff, S. Targeted in situ genome-wide profiling with high efficiency for low cell numbers. Nat. Protoc. 13, 1006–1019 (2018).
Liu, N. et al. Direct promoter repression by BCL11A controls the fetal to adult hemoglobin switch. Cell 173, 430–442 (2018).
Hainer, S. J., Boškovic, A., McCannell, K. N., Rando, O. J. & Fazzio, T. G. Profiling of pluripotency factors in individual stem cells and early embryos. Cell 177, 1319–1329 (2019).
Roth, T. L. et al. Reprogramming human T cell function and specificity with non-viral genome targeting. Nature 559, 405–409 (2018).
Oomen, M. E., Hansen, A. S., Liu, Y., Darzacq, X. & Dekker, J. CTCF sites display cell cycle-dependent dynamics in factor binding and nucleosome positioning. Genome Res. 29, 236–249 (2019).
Zhu, Q., Liu, N., Orkin, S. H. & Yuan, G. C. CUT&RUNTools: a flexible pipeline for CUT&RUN processing and footprint analysis. Genome Biol. 20, 192 (2019).
Meers, M. P., Tenenbaum, D. & Henikoff, S. Peak calling by sparse enrichment analysis for CUT&RUN chromatin profiling. Epigenetics Chromatin 12, 42 (2019).
Meers, M.P., Janssens, D.H. & Henikoff, S. Pioneer factor-nucleosome binding events during differentiation are motif encoded. Mol Cell. 75, 562-575 (2019).
Kaya-Okur, H. S. et al. CUT&Tag for efficient epigenomic profiling of small samples and single cells. Nat. Commun. 10, 1930 (2019).
Harada, A. et al. A chromatin integration labelling method enables epigenomic profiling with lower input. Nat. Cell Biol. 21, 287–296 (2019).
Carter, B. et al. Mapping histone modifications in low cell number and single cells using antibody-guided chromatin tagmentation (ACT-seq). Nat. Commun. 10, 3747 (2019).
Wang, Q. et al. CoBATCH for high-throughput single-cell epigenomic profiling. Mol. Cell 76, 206–216 e7 (2019).
Meers, M. P., Bryson, T. D., Henikoff, J. G. & Henikoff, S. Improved CUT&RUN chromatin profiling tools. Elife 8, e46314 (2019).
Buenrostro, J. D., Giresi, P. G., Zaba, L. C., Chang, H. Y. & Greenleaf, W. J. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218 (2013).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Quinlan, A. R. & Hall, I. M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010).
Ramirez, F. et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44, W160–W165 (2016).
Picelli, S. et al. Tn5 transposase and tagmentation procedures for massively scaled sequencing projects. Genome Res. 24, 2033–2040 (2014).
Buenrostro, J. D. et al. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature 523, 486–490 (2015).
Liu, T. Use model-based Analysis of ChIP-Seq (MACS) to analyze short reads generated by sequencing protein-DNA interactions in embryonic stem cells. Methods Mol. Biol. 1150, 81–95 (2014).
Landt, S. G. et al. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res. 22, 1813–1831 (2012).
Jung, Y. L. et al. Impact of sequencing depth in ChIP-seq experiments. Nucleic Acids Res. 42, e74 (2014).
Oh, K. S., Ha, J., Baek, S. & Sung, M. H. XL-DNase-seq: improved footprinting of dynamic transcription factors. Epigenetics Chromatin 12, 30 (2019).
Ernst, C., Eling, N., Martinez-Jimenez, C. P., Marioni, J. C. & Odom, D. T. Staged developmental mapping and X chromosome transcriptional dynamics during mouse spermatogenesis. Nat. Commun. 10, 1251 (2019).
Org, T. et al. Genome-wide histone modification profiling of inner cell mass and trophectoderm of bovine blastocysts by RAT-ChIP. PLoS ONE 14, e0225801 (2019).
Mo, A. et al. Epigenomic signatures of neuronal diversity in the mammalian brain. Neuron 86, 1369–1384 (2015).
Acknowledgements
We thank Christine Codomo for pooling Illumina sequencing libraries and members of our laboratory and colleagues at the Fred Hutch for providing input. We are especially grateful to the many Protocols.io subscribers around the world who have tried CUT&Tag and provided helpful comments and feedback that have enriched this protocol. This work was supported by the Howard Hughes Medical Institute (H.S.K.-O. and S.H.), grants R01 HG010492 (S.H.) and R01 GM108699 (K.A.) from the National Institutes of Health and an HCA Seed Network grant from the Chan-Zuckerberg Initiative (S.H.).
Author information
Authors and Affiliations
Contributions
H.S.K.-O. and S.H. developed the protocol with input from K.A and D.H.J. S.H. performed the experiments, and with J.G.H. analyzed the data. S.H. and K.A. wrote the manuscript with input from H.S.K.-O, D.H.J., and J.G.H.
Corresponding author
Ethics declarations
Competing interests
H.S.K.-O. and S.H. have filed patent applications related to this work.
Additional information
Peer review information Nature Protocols thanks Sabrina Krueger, Julia Zeitlinger and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key reference using this protocol
Kaya-Okur, H. S. et al. Nat. Commun. 10, 1930 (2019): https://doi.org/10.1038/s41467-019-09982-5
Supplementary information
Supplementary Information
Supplementary Note 1 and Supplementary Fig. 1.
Supplementary Table 1
Primer Sequences.
Rights and permissions
About this article
Cite this article
Kaya-Okur, H.S., Janssens, D.H., Henikoff, J.G. et al. Efficient low-cost chromatin profiling with CUT&Tag. Nat Protoc 15, 3264–3283 (2020). https://doi.org/10.1038/s41596-020-0373-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-020-0373-x
This article is cited by
-
Evaluating histone modification analysis of individual preimplantation embryos
BMC Genomics (2024)
-
Logical design of synthetic cis-regulatory DNA for genetic tracing of cell identities and state changes
Nature Communications (2024)
-
Scalable single-cell profiling of chromatin modifications with sciCUT&Tag
Nature Protocols (2024)
-
Nano-CUT&Tag for multimodal chromatin profiling at single-cell resolution
Nature Protocols (2024)
-
Structure-guided functional suppression of AML-associated DNMT3A hotspot mutations
Nature Communications (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.