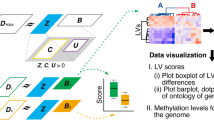
Abstract
DNA methylation profiling offers unique insights into human development and diseases. Often the analysis of complex tissues and cell mixtures is the only feasible option to study methylation changes across large patient cohorts. Since DNA methylomes are highly cell type specific, deconvolution methods can be used to recover cell type–specific information in the form of latent methylation components (LMCs) from such ‘bulk’ samples. Reference-free deconvolution methods retrieve these components without the need for DNA methylation profiles of purified cell types. Currently no integrated and guided procedure is available for data preparation and subsequent interpretation of deconvolution results. Here, we describe a three-stage protocol for reference-free deconvolution of DNA methylation data comprising: (i) data preprocessing, confounder adjustment using independent component analysis (ICA) and feature selection using DecompPipeline, (ii) deconvolution with multiple parameters using MeDeCom, RefFreeCellMix or EDec and (iii) guided biological inference and validation of deconvolution results with the R/Shiny graphical user interface FactorViz. Our protocol simplifies the analysis and guides the initial interpretation of DNA methylation data derived from complex samples. The harmonized approach is particularly useful to dissect and evaluate cell heterogeneity in complex systems such as tumors. We apply the protocol to lung cancer methylomes from The Cancer Genome Atlas (TCGA) and show that our approach identifies the proportions of stromal cells and tumor-infiltrating immune cells, as well as associations of the detected components with clinical parameters. The protocol takes slightly >3 d to complete and requires basic R skills.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
The results shown here are wholly or partially based upon data generated by the TCGA (TCGA-LUAD dataset) Research Network: https://www.cancer.gov/tcga. The Ewing sarcoma dataset is available from the Gene Expression Omnibus GEO, accession number GSE88826.
Code availability
All R packages are available from public code repositories:
DecompPipeline: https://github.com/CompEpigen/DecompPipeline
MeDeCom: https://github.com/lutsik/MeDeCom
FactorViz: https://github.com/CompEpigen/FactorViz
consensusICA: https://gitlab.com/biomodlih/consica.
The pipeline behind our protocol is available as R source packages under open-source licenses (DecompPiepline, MeDeCom, FactorViz: GPLv3; consensusICA: Standard MIT license) and is also implemented as a Docker container available from DockerHub: https://hub.docker.com/r/mscherer/medecom. Supplementary resources and R scripts used to generate the figures are available from http://epigenomics.dkfz.de/DecompProtocol/. The code in this paper has been peer-reviewed.
References
Durek, P. et al. Epigenomic profiling of human CD4+ T cells supports a linear differentiation model and highlights molecular regulators of memory development. Immunity 45, 1148–1161 (2016).
Karpinski, P., Pesz, K. & Sasiadek, M. M. Pan-cancer analysis reveals presence of pronounced DNA methylation drift in CpG island methylator phenotype clusters. Epigenomics 9, 1341–1352 (2017).
Møller, M. et al. Heterogeneous patterns of DNA methylation-based field effects in histologically normal prostate tissue from cancer patients. Sci. Rep. 7, 40636 (2017).
Vidal, E. et al. A DNA methylation map of human cancer at single base-pair resolution. Oncogene 36, 5648–5657 (2017).
Azuara, D. et al. New methylation biomarker panel for early diagnosis of dysplasia or cancer in high-risk inflammatory bowel disease patients. Inflamm. Bowel Dis. 24, 2555–2564 (2018).
Horvath, S. & Raj, K. DNA methylation-based biomarkers and the epigenetic clock theory of ageing. Nat. Rev. Genet. 19, 371–384 (2018).
Stunnenberg, H. G. et al. The International Human Epigenome Consortium: a blueprint for scientific collaboration and discovery. Cell 167, 1145–1149 (2016).
Adams, D. et al. BLUEPRINT to decode the epigenetic signature written in blood. Nat. Biotechnol. 30, 224–226 (2012).
Bock, C. Analysing and interpreting DNA methylation data. Nat. Rev. Genet. 13, 705–719 (2012).
Teschendorff, A. E. & Relton, C. L. Statistical and integrative system-level analysis of DNA methylation data. Nat. Rev. Genet. 19, 129–147 (2017).
Houseman, E. A. et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinforma. 13, 86 (2012).
Teschendorff, A. E., Breeze, C. E., Zheng, S. C. & Beck, S. A comparison of reference-based algorithms for correcting cell-type heterogeneity in Epigenome-Wide Association Studies. BMC Bioinforma. 18, 105 (2017).
Zheng, S. C. et al. A novel cell-type deconvolution algorithm reveals substantial contamination by immune cells in saliva, buccal and cervix. Epigenomics 10, 925–940 (2018).
Chakravarthy, A. et al. Pan-cancer deconvolution of tumour composition using DNA methylation. Nat. Commun. 9, 3220 (2018).
Hicks, S. C. & Irizarry, R. A. methylCC: technology-independent estimation of cell type composition using differentially methylated regions. Genome Biol. 20, 261 (2019).
Salas, L. A. et al. An optimized library for reference-based deconvolution of whole-blood biospecimens assayed using the Illumina HumanMethylationEPIC BeadArray. Genome Biol. 19, 64 (2018).
Kaushal, A. et al. Comparison of different cell type correction methods for genome-scale epigenetics studies. BMC Bioinforma. 18, 216 (2017).
Zou, J., Lippert, C., Heckerman, D., Aryee, M. & Listgarten, J. Epigenome-wide association studies without the need for cell-type composition. Nat. Methods 11, 309–311 (2014).
Rahmani, E. et al. Sparse PCA corrects for cell type heterogeneity in epigenome-wide association studies. Nat. Methods 13, 443–445 (2016).
Rahmani, E. et al. BayesCCE: a Bayesian framework for estimating cell-type composition from DNA methylation without the need for methylation reference. Genome Biol. 19, 141 (2018).
Houseman, E. A. et al. Reference-free deconvolution of DNA methylation data and mediation by cell composition effects. BMC Bioinforma. 17, 259 (2016).
Onuchic, V. et al. Epigenomic deconvolution of breast tumors reveals metabolic coupling between constituent cell types. Cell Rep. 17, 2075–2086 (2016).
Lutsik, P. et al. MeDeCom: discovery and quantification of latent components of heterogeneous methylomes. Genome Biol. 18, 55 (2017).
Rahmani, E. et al. Cell-type-specific resolution epigenetics without the need for cell sorting or single-cell biology. Nat. Commun. 10, 3417 (2019).
Thompson, M., Chen, Z. J., Rahmani, E. & Halperin, E. CONFINED: distinguishing biological from technical sources of variation by leveraging multiple methylation datasets. Genome Biol. 20, 138 (2019).
Decamps, C. et al. Guidelines for cell-type heterogeneity quantification based on a comparative analysis of reference-free DNA methylation deconvolution software. BMC Bioinforma. 21, 16 (2020).
Assenov, Y. et al. Comprehensive analysis of DNA methylation data with RnBeads. Nat. Methods 11, 1138–1140 (2014).
Müller, F. et al. RnBeads 2.0: comprehensive analysis of DNA methylation data. Genome Biol. 20, 55 (2019).
Heyn, H. et al. Distinct DNA methylomes of newborns and centenarians. Proc. Natl Acad. Sci. USA 109, 10522–10527 (2012).
Horvath, S. DNA methylation age of human tissues and cell types. Genome Biol. 14, R115 (2013).
Sompairac, N. et al. Independent component analysis for unraveling the complexity of cancer omics datasets. Int. J. Mol. Sci. 20, 4414 (2019).
Everson, T. M. et al. Cadmium-associated differential methylation throughout the placental genome: epigenome-wide association study of two U.S. birth cohorts. Environ. Health Perspect. 126, 017010 (2018).
Carlström, K. E. et al. Therapeutic efficacy of dimethyl fumarate in relapsing-remitting multiple sclerosis associates with ROS pathway in monocytes. Nat. Commun. 10, 3081 (2019).
Goeppert, B. et al. Integrative analysis defines distinct prognostic subgroups of intrahepatic cholangiocarcinoma. Hepatology 69, 2091–2106 (2019).
Man, Y. G. et al. Tumor-infiltrating immune cells promoting tumor invasion and metastasis: Existing theories. J. Cancer 4, 84–95 (2013).
Reinius, L. E. et al. Differential DNA methylation in purified human blood cells: implications for cell lineage and studies on disease susceptibility. PLoS ONE 7, e41361 (2012).
Luo, C. et al. Single-cell methylomes identify neuronal subtypes and regulatory elements in mammalian cortex. Science 357, 600–604 (2017).
Mulqueen, R. M. et al. Highly scalable generation of DNA methylation profiles in single cells. Nat. Biotechnol. 36, 428–431 (2018).
Lister, R. et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 462, 315–322 (2009).
Meissner, A. et al. Reduced representation bisulfite sequencing for comparative high-resolution DNA methylation analysis. Nucleic Acids Res. 33, 5868–5877 (2005).
Bibikova, M. et al. High density DNA methylation array with single CpG site resolution. Genomics 98, 288–295 (2011).
Aryee, M. J. et al. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics 30, 1363–1369 (2014).
Pidsley, R. et al. A data-driven approach to preprocessing Illumina 450K methylation array data. BMC Genomics 14, 293 (2013).
Tian, Y. et al. ChAMP: updated methylation analysis pipeline for Illumina BeadChips. Bioinformatics 33, 3982–3984 (2017).
Troyanskaya, O. et al. Missing value estimation methods for DNA microarrays. Bioinformatics 17, 520–525 (2001).
Sherry, S. T. et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 29, 308–311 (2001).
Chen, Y. A. et al. Discovery of cross-reactive probes and polymorphic CpGs in the Illumina Infinium HumanMethylation450 microarray. Epigenetics 8, 203–209 (2013).
Pidsley, R. et al. Critical evaluation of the Illumina MethylationEPIC BeadChip microarray for whole-genome DNA methylation profiling. Genome Biol. 17, 208 (2016).
Aran, D., Sirota, M. & Butte, A. J. Systematic pan-cancer analysis of tumour purity. Nat. Commun. 6, 8971 (2015).
Dirkse, A. et al. Stem cell-associated heterogeneity in Glioblastoma results from intrinsic tumor plasticity shaped by the microenvironment. Nat. Commun. 10, 1787 (2019).
Nazarov, P. V. et al. Deconvolution of transcriptomes and miRNomes by independent component analysis provides insights into biological processes and clinical outcomes of melanoma patients. BMC Med. Genomics 12, 132 (2019).
Therneau, T. M. & Grambsch, P. M. Data: Extending the Cox Model (Springer, 2000).
Falcon, S. & Gentleman, R. Using GOstats to test gene lists for GO term association. Bioinformatics 23, 257–258 (2007).
Sheffield, N. C. & Bock, C. LOLA: enrichment analysis for genomic region sets and regulatory elements in R and Bioconductor. Bioinformatics 32, 587–589 (2016).
The Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature 511, 543–550 (2014).
Testa, U., Castelli, G. & Pelosi, E. Lung cancers: molecular characterization, clonal heterogeneity and evolution, and cancer stem cells. Cancers (Basel) 10, 248 (2018).
Teschendorff, A. E. et al. A beta-mixture quantile normalization method for correcting probe design bias in Illumina Infinium 450 k DNA methylation data. Bioinformatics 29, 189–196 (2013).
Cerami, E. et al. The cBio Cancer Genomics Portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401–404 (2012).
Yoshihara, K. et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat. Commun. 4, 2612 (2013).
Travaglini, K. J. et al. A molecular cell atlas of the human lung from single cell RNA sequencing. Preprint at https://www.biorxiv.org/content/10.1101/742320v2 (2020).
Hahn, M. A. et al. Methylation of Polycomb target genes in intestinal cancer is mediated by inflammation. Cancer Res. 68, 10280 (2008).
Varambally, S. et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature 419, 624–629 (2002).
Cai, Y. et al. Epigenetic alterations to Polycomb targets precede malignant transition in a mouse model of breast cancer. Sci. Rep. 8, 5535 (2018).
Ward, M. J. et al. Tumour-infiltrating lymphocytes predict for outcome in HPV-positive oropharyngeal cancer. Br. J. Cancer 110, 489–500 (2014).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2009).
Colaprico, A. et al. TCGAbiolinks: an R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 44, e71 (2016).
Malta, T. M. et al. Machine learning identifies stemness features associated with oncogenic dedifferentiation. Cell 173, 338–354.e15 (2018).
Harris, T. et al. Both gene amplification and allelic loss occur at 14q13.3 in lung cancer. Clin. Cancer Res. 17, 690–699 (2011).
Sheffield, N. C. et al. DNA methylation heterogeneity defines a disease spectrum in Ewing sarcoma. Nat. Med. 23, 386–395 (2017).
Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 57, 289–300 (1995).
The ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012).
Sánchez-Castillo, M. et al. CODEX: a next-generation sequencing experiment database for the haematopoietic and embryonic stem cell communities. Nucleic Acids Res. 43, D1117–D1123 (2015).
Liu, T. et al. Cistrome: an integrative platform for transcriptional regulation studies. Genome Biol. 12, R83 (2011).
Prive, F., Aschard, H., Ziyatdinov, A. & Blum, M. G. B. Efficient analysis of large-scale genome-wide data with two R packages: Bigstatsr and bigsnpr. Bioinformatics 34, 2781–2787 (2018).
Ritchie, M. E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47–e47 (2015).
Houseman, E. A., Molitor, J. & Marsit, C. J. Reference-free cell mixture adjustments in analysis of DNA methylation data. Bioinformatics 30, 1431–1439 (2014).
Jaffe, A. E. & Irizarry, R. A. Accounting for cellular heterogeneity is critical in epigenome-wide association studies. Genome Biol. 15, R31 (2014).
Acknowledgements
We thank the HADACA consortium (Health Data Challenge, Aussois, Dec 2018 and Nov 2019) for valuable input and D. Gupta for thoroughly testing the proposed pipeline. We are grateful to K. Breuer for testing the Docker container, and to F. Azuaje for supporting the collaboration. This work was funded in part by the German Epigenome Project (DEEP, German Science Ministry grant no. 01KU1216A), de.NBI-epi (German Science Ministry grant nos. 031L0101A and 031L0101D) and the EU H2020 project SYSCID (733100). P.V.N. and T.K. were supported by the Luxembourg National Research Fund (C17/BM/11664971/DEMICS). P.L. was supported by the DKFZ Postdoctoral Fellowship and the AMPro Project of the Helmholtz Association (ZT00026).
Author information
Authors and Affiliations
Contributions
M.S. and P.L. implemented most of the computational procedures. P.L. and N.V. previously developed, published and recently updated MeDeCom for installation on Windows. S.S, M.S. and P.L. implemented FactorViz. P.V.N. and T.K. implemented consensus ICA. M.S. performed the analysis of the example datasets, and created all figures and tables. P.V.N., R.T. and V.M. provided crucial input to the analysis and interpretation, and thoroughly tested the protocol. P.L., J.W., T.L. and C.P. jointly supervised the project. M.S. and P.L. wrote the manuscript, with contributions from all co-authors. All authors read and approved the final text.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Protocols thanks Lucas Salas and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key references using this protocol
Lutsik, P. et al. Genome Biol. 18, 55 (2017): https://doi.org/10.1186/s13059-017-1182-6
Müller, F. et al. Genome Biol. 20, 55 (2019): https://doi.org/10.1186/s13059-019-1664-9
Nazarov, P. et al. BMC Med. Genomics 12, 132 (2019): https://doi.org/10.1186/s12920-019-0578-4
Goeppert, B. et al. Hepatology 69, 2091–2106 (2019): https://doi.org/10.1002/hep.30493
Decamps, C. et al. BMC Bioinforma. 21, 16 (2020): https://doi.org/10.1186/s12859-019-3307-2
Key data used in this protocol
The Cancer Genome Atlas Research Network, Nature 511, 543–550 (2014): https://doi.org/10.1038/nature13385
Sheffield, N. et al. Nat. Med. 23, 386–395 (2017): https://doi.org/10.1038/nm.4273
Extended data
Extended Data Fig. 1 Quality control of TCGA data.
a Boxplot for hybridization control probes for the green and the red channel, respectively. Boxplot lines represent the median, the 25th- and 75th- percentiles, and 1.5 times the inter-quartile range. b Sex prediction based on the intensities of the probes on the sex chromosomes. A logistic regression classifier was employed to differentiate between female and male samples. c Outline of the CpG filtering procedure. The sites on the 450k array are filtered according to quality scores (coverage, overall intensity), genomic sequence context (SNPs, sex chromosomes), and cross-reactive sites are discarded.
Extended Data Fig. 2 Selecting the number of components and the regularization parameter for MeDeCom.
a Cross-validation error plotted against the number of latent components K for different values of the regularization parameter λ. Differences across the values of K mask the differences between the five λ values. b Objective value and cross-validation error for different values of λ after fixing the number of components to 7. c Multidimensional scaling of the LMC data matrix after fixing the number of components to 7 and the regularization parameter to 0.001. Shown are the first two multidimensional components. d Violin plots of the LMC methylation matrix for the selected parameters. Boxplot lines represent the median, the 25th- and 75th- percentiles, and 1.5 times the inter-quartile range.
Extended Data Fig. 3 Interpreting RefFreeCellMix results with FactorViz.
a Heatmap of LMC proportions in TCGA-LUAD cohort samples (K=7 components). The samples were hierarchically clustered according to the Euclidean distance between the proportions using complete linkage. We annotated samples using disease status and with the sample-specific LUMP estimate. b Associations between the phenotypic traits and proportions. For quantitative traits, the Pearson correlations are shown as ellipses that are directed to the upper right for positive and to the lower right for negative correlations, respectively. For qualitative traits, the absolute difference of the proportions in the two groups (for example, female vs. male) is shown. P values (two-sided correlation test for quantitative and two-sided t-test for categorical variables) less than 0.01 are indicated by bold borders. LOLA (c) and GO (d) enrichment analysis of the LMC-specific hypomethylated sites for components 1, 2 and 4. No significant GO enrichment was found for components 1 and 4. Sites were defined as LMC-specific hypomethylated if the difference between the value of the methylation component and the median of all other components was less than 0.5. P values have been adjusted for multiple testing with the Benjamini-Hochberg method. e Scatterplots between proportions per sample and known marker gene expression of different lung cell types. The gene expression was measured using counts per million (CPM).
Extended Data Fig. 4 Survival analysis using the survival R-package52 comparing different levels of LMC proportions.
Shown are Kaplan-Meier curves, while samples were stratified according to the LMC proportions into two groups according to the median (high vs. low proportions). P values were computed using the Cox proportional hazards model with the LMC proportions as input, and age, sex, and tumor stage as covariates.
Extended Data Fig. 5 Interpreting MeDeCom results on the Ewing sarcoma RRBS data set69 with FactorViz.
a Heatmap of LMC proportions in the Ewing sarcoma samples (K=6 components, λ=0.001). The samples were hierarchically clustered according to the Euclidean distance between the proportions using complete linkage. We annotated samples using the tumor location and with the sample-specific LUMP estimate. b Associations between the phenotypic traits and proportions. For quantitative traits, the Pearson correlations are shown as ellipses that are directed to the upper right for positive and to the lower right for negative correlations, respectively. For qualitative traits, the absolute difference of the proportions in the two groups (for example mutation vs. wildtype) is shown. P values (two-sided correlation test for quantitative and two-sided t-test for categorical variables) less than 0.01 are indicated by bold borders. GO (c) and LOLA (d) enrichment analysis of the LMC6-specific hypomethylated sites. No significant LOLA and GO enrichments were found for the remaining LMCs. Sites were defined as LMC-specific hypomethylated if the difference between the value of the LMC and the median of all other components was less than 0.5. P values have been adjusted for multiple testing with the Benjamini-Hochberg method. No matched gene-expression values were available for this data set.
Supplementary information
Supplementary Information
Supplementary Note, Supplementary Tables 1–3 and Supplementary Figures 1–8.
Rights and permissions
About this article
Cite this article
Scherer, M., Nazarov, P.V., Toth, R. et al. Reference-free deconvolution, visualization and interpretation of complex DNA methylation data using DecompPipeline, MeDeCom and FactorViz. Nat Protoc 15, 3240–3263 (2020). https://doi.org/10.1038/s41596-020-0369-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-020-0369-6
This article is cited by
-
Club cells employ regeneration mechanisms during lung tumorigenesis
Nature Communications (2022)
-
Tumor fractions deciphered from circulating cell-free DNA methylation for cancer early diagnosis
Nature Communications (2022)
-
Identification of tissue-specific and common methylation quantitative trait loci in healthy individuals using MAGAR
Epigenetics & Chromatin (2021)
-
Machine learning for deciphering cell heterogeneity and gene regulation
Nature Computational Science (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.