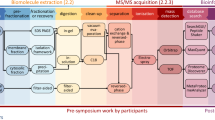
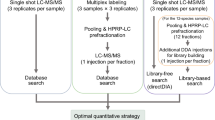
Abstract
Metaproteomics, the study of the collective protein composition of multi-organism systems, provides deep insights into the biodiversity of microbial communities and the complex functional interplay between microbes and their hosts or environment. Thus, metaproteomics has become an indispensable tool in various fields such as microbiology and related medical applications. The computational challenges in the analysis of corresponding datasets differ from those of pure-culture proteomics, e.g., due to the higher complexity of the samples and the larger reference databases demanding specific computing pipelines. Corresponding data analyses usually consist of numerous manual steps that must be closely synchronized. With MetaProteomeAnalyzer and Prophane, we have established two open-source software solutions specifically developed and optimized for metaproteomics. Among other features, peptide-spectrum matching is improved by combining different search engines and, compared to similar tools, metaproteome annotation benefits from the most comprehensive set of available databases (such as NCBI, UniProt, EggNOG, PFAM, and CAZy). The workflow described in this protocol combines both tools and leads the user through the entire data analysis process, including protein database creation, database search, protein grouping and annotation, and results visualization. To the best of our knowledge, this protocol presents the most comprehensive, detailed and flexible guide to metaproteomics data analysis to date. While beginners are provided with robust, easy-to-use, state-of-the-art data analysis in a reasonable time (a few hours, depending on, among other factors, the protein database size and the number of identified peptides and inferred proteins), advanced users benefit from the flexibility and adaptability of the workflow.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
The mass spectrometric datasets analyzed during the current study are available in the PRIDE repository, https://www.ebi.ac.uk/pride/archive/projects/PXD010550. Protein databases, converted mass spectrometry files and example analyses results are available in the Zenodo repository, https://doi.org/10.5281/zenodo.3727600.
Code availability
The code of the tools used in the current study is available in the Zenodo repository (Prophane: https://doi.org/10.5281/zenodo.3727758; MPA: https://doi.org/10.5281/zenodo.3735146) and GitLab (Prophane: https://gitlab.com/s.fuchs/prophane).
References
Gentile, C. L. & Weir, T. L. The gut microbiota at the intersection of diet and human health. Science 362, 776–780 (2018).
Pasolli, E. et al. Extensive unexplored human microbiome diversity revealed by over 150,000 genomes from metagenomes spanning age, geography, and lifestyle. Cell 176, 649–662.e20 (2019).
Routy, B. et al. Gut microbiome influences efficacy of PD-1–based immunotherapy against epithelial tumors. Science 359, 91–97 (2018).
Whelan, F. J. et al. Culture-enriched metagenomic sequencing enables in-depth profiling of the cystic fibrosis lung microbiota. Nat. Microbiol. 5, 379–390 (2020).
Franzosa, E. A. et al. Relating the metatranscriptome and metagenome of the human gut. Proc. Natl Acad. Sci. USA 111, E2329–E2338 (2014).
Benítez-Páez, A., Belda-Ferre, P., Simón-Soro, A. & Mira, A. Microbiota diversity and gene expression dynamics in human oral biofilms. BMC Genomics 15, 311 (2014).
Kleiner, M. et al. Assessing species biomass contributions in microbial communities via metaproteomics. Nat. Commun. 8, 1558 (2017).
Cerdó, T. et al. Gut microbial functional maturation and succession during human early life. Environ. Microbiol. 20, 2160–2177 (2018).
Chevrette, M. G. et al. The antimicrobial potential of Streptomyces from insect microbiomes. Nat. Commun. 10, 516 (2019).
Kleiner, M. Metaproteomics: much more than measuring gene expression in microbial communities. mSystems 4, e00115–e00119 (2019).
Ram, R. J. et al. Community proteomics of a natural microbial biofilm. Science 308, 1915–1920 (2005).
Erickson, A. R. et al. Integrated metagenomics/metaproteomics reveals human host-microbiota signatures of Crohn’s disease. PloS ONE 7, e49138 (2012).
Kolmeder, C. A. et al. Colonic metaproteomic signatures of active bacteria and the host in obesity. Proteomics 15, 3544–3552 (2015).
Lum, K. K. & Cristea, I. M. Proteomic approaches to uncovering virus-host protein interactions during the progression of viral infection. Expert Rev. Proteom. 13, 325–340 (2016).
Rabe, A. et al. Metaproteomics analysis of microbial diversity of human saliva and tongue dorsum in young healthy individuals. J. Oral. Microbiol. 11, 1654786 (2019).
Lamont, E. A. et al. Circulating Mycobacterium bovis peptides and host response proteins as biomarkers for unambiguous detection of subclinical infection. J. Clin. Microbiol. 52, 536–543 (2014).
Hettich, R. L., Pan, C., Chourey, K. & Giannone, R. J. Metaproteomics: Harnessing the power of high performance mass spectrometry to identify the suite of proteins that control metabolic activities in microbial communities. Anal. Chem. 85, 4203–4214 (2013).
Muth, T. et al. The MetaProteomeAnalyzer: a powerful open-source software suite for metaproteomics data analysis and interpretation. J. Proteome Res. 14, 1557–1565 (2015).
Perkins, D. N., Pappin, D. J. C., Creasy, D. M. & Cottrell, J. S. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20, 3551–3567 (1999).
Mehlan, H. et al. Data visualization in environmental proteomics. Proteomics 13, 2805–2821 (2013).
Muth, T. et al. MPA portable: a stand-alone software package for analyzing metaproteome samples on the go. Anal. Chem. 90, 685–689 (2018).
Schneider, T. et al. Structure and function of the symbiosis partners of the lung lichen (Lobaria pulmonaria L. Hoffm.) analyzed by metaproteomics. Proteomics 11, 2752–2756 (2011).
Grube, M. et al. Exploring functional contexts of symbiotic sustain within lichen-associated bacteria by comparative omics. ISME J. 9, 412–424 (2015).
Daims, H. et al. Complete nitrification by Nitrospira bacteria. Nature 528, 504–509 (2015).
Eymann, C. et al. Symbiotic interplay of fungi, algae, and bacteria within the lung lichen lobaria pulmonaria L. Hoffm. as assessed by state-of-the-art metaproteomics. J. Proteome Res. 16, 2160–2173 (2017).
Cernava, T. et al. Deciphering functional diversification within the lichen microbiota by meta-omics. Microbiome 5, 82 (2017).
Lassek, C. et al. A metaproteomics approach to elucidate host and pathogen protein expression during catheter-associated urinary tract infections (CAUTIs). Mol. Cell. Proteom. 14, 989–1008 (2015).
Keiblinger, K. M., Fuchs, S., Zechmeister-Boltenstern, S. & Riedel, K. Soil and leaf litter metaproteomics—a brief guideline from sampling to understanding. FEMS Microbiol. Ecol. 92, fiw180 (2016).
Heyer, R. et al. A robust and universal metaproteomics workflow for research studies and routine diagnostics within 24 h using phenol extraction, FASP digest, and the MetaProteomeAnalyzer. Front. Microbiol. 10, 1883 (2019).
Schiebenhoefer, H. et al. Challenges and promise at the interface of metaproteomics and genomics: an overview of recent progress in metaproteogenomic data analysis. Expert Rev. Proteom. 16, 375–390 (2019).
Muth, T., Renard, B. Y. & Martens, L. Metaproteomic data analysis at a glance: advances in computational microbial community proteomics. Expert Rev. Proteom. 13, 757–769 (2016).
Heyer, R. et al. Challenges and perspectives of metaproteomic data analysis. J. Biotechnol. 261, 24–36 (2017).
Wilmes, P. & Bond, P. L. The application of two-dimensional polyacrylamide gel electrophoresis and downstream analyses to a mixed community of prokaryotic microorganisms. Environ. Microbiol. 6, 911–920 (2004).
Blakeley, P., Overton, I. M. & Hubbard, S. J. Addressing statistical biases in nucleotide-derived protein databases for proteogenomic search strategies. J. Proteome Res. 11, 5221–5234 (2012).
Muth, T. et al. Navigating through metaproteomics data: a logbook of database searching. Proteomics 15, 3439–3453 (2015).
Nesvizhskii, A. I. & Aebersold, R. Interpretation of shotgun proteomic data: the protein inference problem. Mol. Cell. Proteom. 4, 1419–1440 (2005).
Nesvizhskii, A. I. Proteogenomics: concepts, applications and computational strategies. Nat. Methods 11, 1114–1125 (2014).
Abraham, P. E., Giannone, R. J., Xiong, W. & Hettich, R. L. Metaproteomics: extracting and mining proteome information to characterize metabolic activities in microbial communities. Curr. Protoc. Bioinformatics 46, 13.26.1–13.26.14 (2014).
Barsnes, H. & Vaudel, M. SearchGUI: a highly adaptable common interface for proteomics search and de novo engines. J. Proteome Res. 17, 2552–2555 (2018).
Vaudel, M. et al. PeptideShaker enables reanalysis of MS-derived proteomics data sets. Nat. Biotechnol. 33, 22–24 (2015).
Tyanova, S., Temu, T. & Cox, J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 11, 2301–2319 (2016).
Boekel, J. et al. Multi-omic data analysis using Galaxy. Nat. Biotechnol. 33, 137–139 (2015).
Blank, C. et al. Disseminating metaproteomic informatics capabilities and knowledge using the galaxy-P framework. Proteomes 6, 7 (2018).
Sachsenberg, T. et al. MetaProSIP: automated inference of stable isotope incorporation rates in proteins for functional metaproteomics. J. Proteome Res. 14, 619–627 (2015).
Gurdeep Singh, R. et al. Unipept 4.0: functional analysis of metaproteome data. J. Proteome Res. 18, 606–615 (2019).
Werner, J., Géron, A., Kerssemakers, J. & Matallana-Surget, S. mPies: a novel metaproteomics tool for the creation of relevant protein databases and automatized protein annotation. Biol. Direct 14, 21 (2019).
Riffle, M. et al. MetaGOmics: a web-based tool for peptide-centric functional and taxonomic analysis of metaproteomics data. Proteomes 6, 2 (2017).
Cheng, K. et al. MetaLab: an automated pipeline for metaproteomic data analysis. Microbiome 5, 157 (2017).
Liao, B. et al. iMetaLab 1.0: a web platform for metaproteomics data analysis. Bioinformatics 34, 3954–3956 (2018).
Vishwanath, S., de Brevern, A. G. & Srinivasan, N. Same but not alike: structure, flexibility and energetics of domains in multi-domain proteins are influenced by the presence of other domains. PLOS Comput. Biol. 14, e1006008 (2018).
Tanca, A. et al. The impact of sequence database choice on metaproteomic results in gut microbiota studies. Microbiome 4, 51 (2016).
Tanca, A. et al. Evaluating the impact of different sequence databases on metaproteome analysis: insights from a lab-assembled microbial mixture. PLoS ONE 8, e82981 (2013).
Timmins-Schiffman, E. et al. Critical decisions in metaproteomics: achieving high confidence protein annotations in a sea of unknowns. ISME J. 11, 309–314 (2017).
Choi, H. & Nesvizhskii, A. I. False discovery rates and related statistical concepts in mass spectrometry-based proteomics. J. Proteome Res. 7, 47–50 (2008).
Kuhring, M. & Renard, B. Y. Estimating the computational limits of detection of microbial non-model organisms. Proteomics 15, 3580–3584 (2015).
Rho, M., Tang, H. & Ye, Y. FragGeneScan: predicting genes in short and error-prone reads. Nucleic Acids Res. 38, e191–e191 (2010).
Tang, H., Li, S. & Ye, Y. A graph-centric approach for metagenome-guided peptide and protein identification in metaproteomics. PLOS Comput. Biol. 12, e1005224 (2016).
Human Microbiome Project Consortium. A framework for human microbiome research. Nature 486, 215–221 (2012).
Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214 (2012).
Dewhirst, F. E. et al. The human oral microbiome. J. Bacteriol. 192, 5002–5017 (2010).
Sunagawa, S. et al. Structure and function of the global ocean microbiome. Science 348, 1261359 (2015).
Liolios, K. et al. The Genomes On Line Database (GOLD) in 2009: status of genomic and metagenomic projects and their associated metadata. Nucleic Acids Res. 38, D346–D354 (2010).
The UniProt Consortium. UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 47, D506–D515 (2019).
O’Leary, N. A. et al. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 44, D733–D745 (2016).
Chambers, M. C. et al. A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol. 30, 918–920 (2012).
Craig, R. & Beavis, R. C. TANDEM: matching proteins with tandem mass spectra. Bioinformatics 20, 1466–1467 (2004).
Geer, L. Y. et al. Open mass spectrometry search algorithm. J. Proteome Res. 3, 958–964 (2004).
Ondov, B. D., Bergman, N. H. & Phillippy, A. M. Interactive metagenomic visualization in a Web browser. BMC Bioinforma. 12, 385 (2011).
Hulstaert, N. et al. ThermoRawFileParser: modular, scalable, and cross-platform RAW file conversion. J. Proteome Res. 19, 537–542 (2020).
Heyer, R. et al. Metaproteome analysis of the microbial communities in agricultural biogas plants. N. Biotechnol. 30, 614–622 (2013).
Heyer, R. et al. Proteotyping of biogas plant microbiomes separates biogas plants according to process temperature and reactor type. Biotechnol. Biofuels 9, 155 (2016).
Buchfink, B., Xie, C. & Huson, D. H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 12, 59–60 (2015).
Huerta-Cepas, J. et al. Fast genome-wide functional annotation through orthology assignment by eggNOG-Mapper. Mol. Biol. Evol. 34, 2115–2122 (2017).
Huerta-Cepas, J. et al. eggNOG 4.5: a hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res. 44, D286–D293 (2016).
Eddy, S. R. Accelerated profile HMM searches. PLOS Comput. Biol. 7, e1002195 (2011).
El-Gebali, S. et al. The Pfam protein families database in 2019. Nucleic Acids Res. 47, D427–D432 (2019).
Haft, D. H. et al. TIGRFAMs and genome properties in 2013. Nucleic Acids Res. 41, D387–D395 (2013).
Zhang, H. et al. dbCAN2: a meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 46, W95–W101 (2018).
Prestat, E. et al. FOAM (Functional Ontology Assignments for Metagenomes): a Hidden Markov Model (HMM) database with environmental focus. Nucleic Acids Res. 42, e145–e145 (2014).
Gibson, M. K., Forsberg, K. J. & Dantas, G. Improved annotation of antibiotic resistance determinants reveals microbial resistomes cluster by ecology. ISME J. 9, 207–216 (2015).
Katoh, K. & Standley, D. M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013).
Elias, J. E. & Gygi, S. P. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 4, 207–214 (2007).
Huson, D. H., Auch, A. F., Qi, J. & Schuster, S. C. MEGAN analysis of metagenomic data. Genome Res. 17, 377–386 (2007).
Zybailov, B. et al. Statistical analysis of membrane proteome expression changes in Saccharomyces cerevisiae. J. Proteome Res. 5, 2339–2347 (2006).
Audain, E. et al. In-depth analysis of protein inference algorithms using multiple search engines and well-defined metrics. J. Proteom. 150, 170–182 (2017).
Deutsch, E. W. et al. Expanding the use of spectral libraries in proteomics. J. Proteome Res. 17, 4051–4060 (2018).
Muth, T., Hartkopf, F., Vaudel, M. & Renard, B. Y. A potential golden age to come-current tools, recent use cases, and future avenues for de novo sequencing in proteomics. Proteomics 18, e1700150 (2018).
Yates, J. R., Eng, J. K., McCormack, A. L. & Schieltz, D. Method to correlate tandem mass spectra of modified peptides to amino acid sequences in the protein database. Anal. Chem. 67, 1426–1436 (1995).
Eng, J. K., Jahan, T. A. & Hoopmann, M. R. Comet: An open-source MS/MS sequence database search tool. Proteomics 13, 22–24 (2013).
Kim, S. & Pevzner, P. A. MS-GF+ makes progress towards a universal database search tool for proteomics. Nat. Commun. 5, 1–10 (2014).
Cox, J. et al. Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10, 1794–1805 (2011).
Park, S. K. R. et al. ComPIL 2.0: an updated comprehensive metaproteomics database. J. Proteome Res. 18, 616–622 (2019).
Beyter, D., Lin, M. S., Yu, Y., Pieper, R. & Bafna, V. ProteoStorm: an ultrafast metaproteomics database search framework. Cell Syst. 7, 463–467.e6 (2018).
Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990).
Henikoff, S. & Henikoff, J. G. Amino acid substitution matrices from protein blocks. Proc. Natl Acad. Sci. USA 89, 10915–10919 (1992).
Acknowledgements
We are grateful to D. Zühlke and L. Gierse for suggesting the importance of Prophane and reporting bugs, F. Hartkopf and X. Wang for carefully evaluating this protocol, and D. Micheel and R. Zoun for helping with the Prophane web service. This project has been supported by the Deutsche Forschungsgemeinschaft (DFG; grant numbers RE3474/5-1 and RE3474/2-2), the de.NBI network (MetaProtServ de-NBI-039), and the BMBF-funded de.NBI Cloud within the German Network for Bioinformatics Infrastructure (de.NBI; 031A537B, 031A533A, 031A538A, 031A533B, 031A535A, 031A537C, 031A534A and 031A532B).
Author information
Authors and Affiliations
Contributions
Workflow design: Steps 1−13, H.S., B.R., K.S., E.S., T.M., K.R. and S.F.; Steps 14−43, K.S., T.M. and D.B.; Steps 44−53, H.S., B.R., K.S., E.S., T.M., K.R. and S.F.; Step 54, H.S., K.S., T.M. and S.F.; training data: K.S. and D.B.; protocol testing: H.S., K.S. and S.F.; manuscript writing: all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key references using this protocol
Lassek, C. et al. Mol. Cell. Proteomics 14, 989–1008 (2015): https://www.mcponline.org/content/early/2015/02/11/mcp.M114.043463
Muth, T. et al. J. Proteome Res. 6, 14:1557–1565 (2015): https://pubs.acs.org/doi/10.1021/pr501246w
Grube, M. et al. ISME J. 9, 412–424 (2015): https://www.nature.com/articles/ismej2014138
Supplementary information
Rights and permissions
About this article
Cite this article
Schiebenhoefer, H., Schallert, K., Renard, B.Y. et al. A complete and flexible workflow for metaproteomics data analysis based on MetaProteomeAnalyzer and Prophane. Nat Protoc 15, 3212–3239 (2020). https://doi.org/10.1038/s41596-020-0368-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-020-0368-7
This article is cited by
-
Ecosystem-specific microbiota and microbiome databases in the era of big data
Environmental Microbiome (2022)
-
Gut microbiome alterations and gut barrier dysfunction are associated with host immune homeostasis in COVID-19 patients
BMC Medicine (2022)
-
Associated bacterial microbiome responds opportunistic once algal host Scenedesmus vacuolatus is attacked by endoparasite Amoeboaphelidium protococcarum
Scientific Reports (2022)
-
Physiological response and proteomics analysis of Reaumuria soongorica under salt stress
Scientific Reports (2022)
-
Gut microbiota as an antioxidant system in centenarians associated with high antioxidant activities of gut-resident Lactobacillus
npj Biofilms and Microbiomes (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.