Abstract
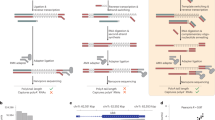
Several noncanonical initial nucleotides (NCINs) have been found to cap RNAs and possibly regulate RNA stability, transcription and translation. NAD+ is one of the NCINs that has recently been discovered to cap RNAs in a wide range of species. Identification of the NAD+-capped RNAs (NAD-RNAs) could help to unveil the cap-mediated regulation mechanisms. We previously reported a method termed NAD tagSeq for genome-wide analysis of NAD-RNAs. NAD tagSeq is based on the previously published NAD captureSeq protocol, which applies an enzymatic reaction and a click chemistry reaction to label NAD-RNAs with biotin followed by enrichment with streptavidin resin and identification by RNA sequencing. In the current NAD tagSeq method, NAD-RNAs are labeled with a synthetic RNA tag and identified by direct RNA sequencing based on Oxford Nanopore technology. Compared to NAD captureSeq, NAD tagSeq provides a simpler procedure for direct sequencing of NAD-RNAs and noncapped RNAs and affords information on the whole sequence organization of NAD-RNAs and the ratio of NAD-RNAs to total transcripts. Furthermore, NAD-RNAs can be enriched by hybridizing a complementary DNA probe to the RNA tag, thus increasing the sequencing coverage of NAD-RNAs. The strategy of tagging RNAs with a synthetic RNA tag and identifying them by direct RNA sequencing might be employed in analyzing other NCIN-capped RNAs. The experimental procedure of NAD tagSeq, including RNA extraction, RNA tagging and direct RNA sequencing, takes ~5 d, and initial data analysis can be completed within 2 d.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The data sets analyzed with the current protocol are available in the Gene Expression Omnibus repository under the accession number GSE127755. The web links for the raw data are https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE127755, https://www.ebi.ac.uk/ena/data/view/PRJNA525282 and https://github.com/dorothyzh/TagSeqTools.
Code availability
All the software used in this protocol, including our Python script for sorting tagged and untagged RNA, are available in the ‘TagSeqTools’ repository under the Apache License v2.0 (https://github.com/dorothyzh/TagSeqTools) and as Supplementary Software.
References
Padgett, R. A. New roles for old RNA cap flavors. Nat. Chem. Biol. 15, 317–318 (2019).
Shuman, S. What messenger RNA capping tells us about eukaryotic evolution. Nat. Rev. Mol. Cell Biol. 3, 619–625 (2002).
Ramanathan, A., Robb, G. B. & Chan, S. H. mRNA capping: biological functions and applications. Nucleic Acids Res. 44, 7511–7526 (2016).
Kwasnik, A. et al. Arabidopsis DXO1 links RNA turnover and chloroplast function independently of its enzymatic activity. Nucleic Acids Res. 47, 4751–4764 (2019).
Wang, Y. et al. NAD+-capped RNAs are widespread in the Arabidopsis transcriptome and can probably be translated. Proc. Natl Acad. Sci. USA 116, 12094–12102 (2019).
Bird, J. G. et al. Highly efficient 5′ capping of mitochondrial RNA with NAD+ and NADH by yeast and human mitochondrial RNA polymerase. eLife 7, e42179 (2018).
Bird, J. G. et al. The mechanism of RNA 5′ capping with NAD+, NADH and desphospho-CoA. Nature 535, 444–447 (2016).
Julius, C. & Yuzenkova, Y. Bacterial RNA polymerase caps RNA with various cofactors and cell wall precursors. Nucleic Acids Res. 45, 8282–8290 (2017).
Grudzien-Nogalska, E., Bird, J. G., Nickels, B. E. & Kiledjian, M. “NAD-capQ” detection and quantitation of NAD caps. RNA 24, 1418–1425 (2018).
Verdin, E. NAD+ in aging, metabolism, and neurodegeneration. Science 350, 1208–1213 (2015).
Grudzien-Nogalska, E. et al. Structural and mechanistic basis of mammalian Nudt12 RNA deNADding. Nat. Chem. Biol. 15, 575–582 (2019).
Winz, M. L. et al. Capture and sequencing of NAD-capped RNA sequences with NAD captureSeq. Nat. Protoc. 12, 122–149 (2017).
Frindert, J. et al. Identification, biosynthesis, and decapping of NAD-capped RNAs in B. subtilis. Cell Rep. 24, 1890–1901 (2018).
Walters, R. W. et al. Identification of NAD+ capped mRNAs in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA 114, 480–485 (2017).
Zhang, H. et al. NAD tagSeq reveals that NAD+-capped RNAs are mostly produced from a large number of protein-coding genes in Arabidopsis. Proc. Natl Acad. Sci. USA 116, 12072–12077 (2019).
Jiao, X. et al. 5′ end nicotinamide adenine dinucleotide cap in human cells promotes RNA decay through DXO-mediated deNADding. Cell 168, 1015–1027 (2017).
Vvedenskaya, I. O. et al. CapZyme-Seq comprehensively defines promoter-sequence determinants for RNA 5’ capping with NAD+. Mol. Cell 70, 553––564 (2018).
Jiang, F. et al. Long-read direct RNA sequencing by 5′-Cap capturing reveals the impact of Piwi on the widespread exonization of transposable elements in locusts. RNA Biol. 16, 950–959 (2019).
Garalde, D. R. et al. Highly parallel direct RNA sequencing on an array of nanopores. Nat. Methods 15, 201–206 (2018).
Wick, R. R., Judd, L. M. & Holt, K. E. Performance of neural network basecalling tools for Oxford Nanopore sequencing. Genome Biol. 20, 129 (2019).
Robinson, J. T. et al. Integrative genomics viewer. Nat. Biotechnol. 29, 24–26 (2011).
Depledge, D. P. et al. Direct RNA sequencing on nanopore arrays redefines the transcriptional complexity of a viral pathogen. Nat. Commun. 10, 754 (2019).
Mauer, J. et al. Reversible methylation of m6Am in the 5′ cap controls mRNA stability. Nature 541, 371–375 (2017).
Warminski, M., Sikorski, P. J., Kowalska, J. & Jemielity, J. Applications of phosphate modification and labeling to study (m)RNA caps. Top. Curr. Chem. 375, 16 (2017).
Kowtoniuk, W. E., Shen, Y., Heemstra, J. M., Agarwal, I. & Liu, D. R. A chemical screen for biological small molecule-RNA conjugates reveals CoA-linked RNA. Proc. Natl Acad. Sci. USA 106, 7768–7773 (2009).
Chen, Y. G., Kowtoniuk, W. E., Agarwal, I., Shen, Y. & Liu, D. R. LC/MS analysis of cellular RNA reveals NAD-linked RNA. Nat. Chem. Biol. 5, 879–881 (2009).
Akichika, S. et al. Cap-specific terminal N6-methylation of RNA by an RNA polymerase II-associated methyltransferase. Science 363, eaav0080 (2019).
Beverly, M., Dell, A., Parmar, P. & Houghton, L. Label-free analysis of mRNA capping efficiency using RNase H probes and LC–MS. Anal. Bioanal. Chem. 408, 5021–5030 (2016).
Wang, J. et al. Quantifying the RNA cap epitranscriptome reveals novel caps in cellular and viral RNA. Nucleic Acids Res. 47, e130 (2019).
Schwartz, S. & Motorin, Y. Next-generation sequencing technologies for detection of modified nucleotides in RNAs. RNA Biol. 14, 1124–1137 (2017).
Cell RepBenjamini, Y. & Speed, T. P. Summarizing and correcting the GC content bias in high-throughput sequencing. Nucleic Acids Res. 40, e72 (2012).
Vallejos, C. A., Risso, D., Scialdone, A., Dudoit, S. & Marioni, J. C. Normalizing single-cell RNA sequencing data: challenges and opportunities. Nat. Methods 14, 565–571 (2017).
Soneson, C. et al. A comprehensive examination of Nanopore native RNA sequencing for characterization of complex transcriptomes. Nat. Commun. 10, 3359 (2019).
Workman, R. E. et al. Nanopore native RNA sequencing of a human poly(A) transcriptome. Nat. Methods 16, 1297–1305 (2019).
Shimizu, Y. et al. Cell-free translation reconstituted with purified components. Nat. Biotechnol. 19, 751–755 (2001).
Johnson, M. RNA extraction. Mater. Methods 2, 201 (2012).
Li, H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34, 3094–3100 (2018).
Zhong, H., Cai, Z., Yang, Z. & Xia, Y. TagSeqTools: a flexible and comprehensive analysis pipeline for NAD tagSeq data. Preprint at https://www.biorxiv.org/content/10.1101/2020.03.09.982934v1 (2020).
Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data (Babraham Bioinformatics, 2010).
Acknowledgements
This work was supported by the National Natural Science Foundation of China (91543202 and 21705137 to Z.C.), Hong Kong Baptist University (SDF 15-1012-P04 and RC-ICRS/16-17/04 to Y.X. and Z.C.), the National Key R&D Program of China (2017YFC1600500 to Z.C.) and the Research Grants Council of Hong Kong (GRF grant nos. 12100415, 12100018, 12100717, 12102719, C2009-19GF and AoE/M-403/16 to Y.X.).
Author information
Authors and Affiliations
Contributions
All authors contributed to the design of the experiments; H. Zhang and X.S. performed experiments; H. Zhong and Z.Y. performed data analysis; all authors analyzed experimental results and contributed to the writing of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Protocols thanks Shobbir Hussain and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Related links
Key reference using this protocol
Zhang, H. et al. Proc. Natl Acad. Sci. USA 116, 12072–12077 (2019): https://doi.org/10.1073/pnas.1903683116
This protocol is an extension to Nat. Protoc. 12, 122–149 (2017): https://doi.org/10.1038/nprot.2016.163
Supplementary information
Supplementary Information
Supplementary Methods and Supplementary Fig. 1.
Supplementary Software
Software for processing NAD tagSeq data and demo files.
Rights and permissions
About this article
Cite this article
Shao, X., Zhang, H., Yang, Z. et al. NAD tagSeq for transcriptome-wide identification and characterization of NAD+-capped RNAs. Nat Protoc 15, 2813–2836 (2020). https://doi.org/10.1038/s41596-020-0363-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-020-0363-z
This article is cited by
-
Arabidopsis DXO1 activates RNMT1 to methylate the mRNA guanosine cap
Nature Communications (2023)
-
A systems-level mass spectrometry-based technique for accurate and sensitive quantification of the RNA cap epitranscriptome
Nature Protocols (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.