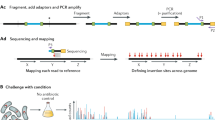
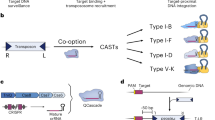
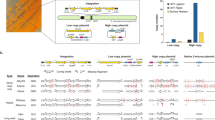
Abstract
Invasive fungal infections caused by Candida species are life threatening with high mortality, posing a severe public health threat. New technologies for rapid, genome-wide identification of virulence genes and therapeutic targets are urgently needed. Our recent engineering of a piggyBac (PB) transposon-mediated mutagenesis system in haploid Candida albicans provides a powerful discovery tool, which we anticipate should be adaptable to other haploid Candida species. In this protocol, we use haploid C. albicans as an example to present an improved version of the mutagenesis system and provide a detailed description of the protocol for constructing high-quality mutant libraries. We also describe a method for quantitative PB insertion site sequencing, PBISeq. The PBISeq library preparation procedure exploits tagmentation to quickly and efficiently construct sequencing libraries. Finally, we present a pipeline to analyze PB insertion sites in a de novo assembled genome of our engineered haploid C. albicans strain. The entire protocol takes ~7 d from transposition induction to having a final library ready for sequencing. This protocol is highly efficient and less labor intensive than alternative approaches and significantly accelerates genetic studies of Candida.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
Demo data derived from using a freshly induced transposon mutant library has been deposited in the NCBI Sequence Read Archive under accession numbers SRX6817047, SRX6817048 and SRX6817049 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA564479). The haploid C. albicans GZY892 assembly and annotation reported here are available as Supplementary Data 1. Raw sequence reads from the Illumina library and the PacBio library have been deposited in NCBI under accession numbers PRJNA605578 and PRJNA605577, respectively. This Whole Genome Shotgun project has also been deposited at DDBJ/ENA/GenBank under the accession number JAAGWN000000000. The version described in this paper is the first version, JAAGWN010000000.
Code availability
The in-house scripts of PBISeq are publicly available in GitHub at https://github.com/xchromosome219/PBseq.pipeline under an MIT license.
References
Nash, A. K. et al. The gut mycobiome of the Human Microbiome Project healthy cohort. Microbiome 5, 153 (2017).
Ghannoum, M. A. et al. Characterization of the oral fungal microbiome (mycobiome) in healthy individuals. PLoS Pathog. 6, e1000713 (2010).
Kam, A. P. & Xu, J. Diversity of commensal yeasts within and among healthy hosts. Diagn. Microbiol. Infect. Dis. 43, 19–28 (2002).
Khullar, G. et al. Chronic mucocutaneous candidiasis. J. Allergy Clin. Immunol. Pract. 5, 1119–1121 (2017).
Brown, G. D. et al. Hidden killers: human fungal infections. Sci. Transl. Med. 4, 165rv113 (2012).
Bodey, G. P. The emergence of fungi as major hospital pathogens. J. Hosp. Infect. 11(Suppl A), 411–426 (1988).
Groll, A. H., Piscitelli, S. C. & Walsh, T. J. Clinical pharmacology of systemic antifungal agents: a comprehensive review of agents in clinical use, current investigational compounds, and putative targets for antifungal drug development. Adv. Pharmacol. 44, 343–500 (1998).
Georgopapadakou, N. H. & Walsh, T. J. Human mycoses: drugs and targets for emerging pathogens. Science 264, 371–373 (1994).
Hickman, M. A. et al. The ‘obligate diploid’ Candida albicans forms mating-competent haploids. Nature 494, 55–59 (2013).
Seneviratne, C. J. et al. New “haploid biofilm model” unravels IRA2 as a novel regulator of Candida albicans biofilm formation. Sci. Rep. 5, 12433 (2015).
Yang, S. L. et al. Sac7 and Rho1 regulate the white-to-opaque switching in Candida albicans. Sci. Rep. 8, 875 (2018).
Huang, Z. X., Wang, H., Wang, Y. M. & Wang, Y. Novel mechanism coupling cyclic AMP-protein kinase A signaling and Golgi trafficking via Gyp1 phosphorylation in polarized growth. Eukaryot. Cell 13, 1548–1556 (2014).
Gao, J. et al. Candida albicans gains azole resistance by altering sphingolipid composition. Nat. Commun. 9, 4495 (2018).
Shapiro, R. S. et al. A CRISPR-Cas9-based gene drive platform for genetic interaction analysis in Candida albicans. Nat. Microbiol. 3, 73–82 (2018).
Yusa, K. piggyBac Transposon. Microbiol. Spectr. 3, 2 (2015).
Cary, L. C. et al. Transposon mutagenesis of baculoviruses: analysis of Trichoplusia ni transposon IFP2 insertions within the FP-locus of nuclear polyhedrosis viruses. Virology 172, 156–169 (1989).
Ding, S. et al. Efficient transposition of the piggyBac (PB) transposon in mammalian cells and mice. Cell 122, 473–483 (2005).
Mitra, R., Fain-Thornton, J. & Craig, N. L. piggyBac can bypass DNA synthesis during cut and paste transposition. Embo J. 27, 1097–1109 (2008).
Segal, E. S. et al. Gene essentiality analyzed by in vivo transposon mutagenesis and machine learning in a stable haploid isolate of Candida albicans. MBio 9, e02048-18 (2018).
Rodrigues, C. F., Silva, S. & Henriques, M. Candida glabrata: a review of its features and resistance. Eur. J. Clin. Microbiol. Infect. Dis. 33, 673–688 (2014).
de Cassia Orlandi Sardi, J., Silva, D. R., Soares Mendes-Giannini, M. J. & Rosalen, P. L. Candida auris: epidemiology, risk factors, virulence, resistance, and therapeutic options. Microb. Pathog. 125, 116–121 (2018).
Mielich, K. et al. Maize transposable elements Ac/Ds as insertion mutagenesis tools in Candida albicans. G3 (Bethesda) 8, 1139–1145 (2018).
Yusa, K. et al. A hyperactive piggyBac transposase for mammalian applications. Proc. Natl Acad. Sci. USA 108, 1531–1536 (2011).
Devon, R. S., Porteous, D. J. & Brookes, A. J. Splinkerettes-improved vectorettes for greater efficiency in PCR walking. Nucleic Acids Res. 23, 1644–1645 (1995).
Bronner, I. F. et al. Quantitative insertion-site sequencing (QIseq) for high throughput phenotyping of transposon mutants. Genome Res. 26, 980–989 (2016).
Seguin-Orlando, A. et al. Ligation bias in Illumina next-generation DNA libraries: implications for sequencing ancient genomes. PLoS ONE 29, e78575 (2013).
Uhl, M. A., Biery, M., Craig, N. & Johnson, A. D. Haploinsufficiency-based large-scale forward genetic analysis of filamentous growth in the diploid human fungal pathogen C. albicans. Embo J. 22, 2668–2678 (2003).
Davis, D. A., Bruno, V. M., Loza, L., Filler, S. G. & Mitchell, A. P. Candida albicans Mds3p, a conserved regulator of pH responses and virulence identified through insertional mutagenesis. Genetics 162, 1573–1581 (2002).
Noble, S. M., French, S., Kohn, L. A., Chen, V. & Johnson, A. D. Systematic screens of a Candida albicans homozygous deletion library decouple morphogenetic switching and pathogenicity. Nat. Genet. 42, 590–598 (2010).
Bharucha, N. et al. A large-scale complex haploinsufficiency-based genetic interaction screen in Candida albicans: analysis of the RAM network during morphogenesis. PLoS Genet. 7, e1002058 (2011).
Fuller, K. K., Chen, S., Loros, J. J. & Dunlap, J. C. Development of the CRISPR/Cas9 system for targeted gene disruption in Aspergillus fumigatus. Eukaryot. Cell 14, 1073–1080 (2015).
Liu, R., Chen, L., Jiang, Y., Zhou, Z. & Zou, G. Efficient genome editing in filamentous fungus Trichoderma reesei using the CRISPR/Cas9 system. Cell Discov. 1, 15007 (2015).
Vyas, V. K., Barrasa, M. I. & Fink, G. R. A Candida albicans CRISPR system permits genetic engineering of essential genes and gene families. Sci. Adv. 1, e1500248 (2015).
Enkler, L., Richer, D., Marchand, A. L., Ferrandon, D. & Jossinet, F. Genome engineering in the yeast pathogen Candida glabrata using the CRISPR-Cas9 system. Sci. Rep. 6, 35766 (2016).
Min, K., Ichikawa, Y., Woolford, C. A. & Mitchell, A. P. Candida albicans gene deletion with a transient CRISPR-Cas9 system. mSphere 1, e00130-16 (2016).
Grahl, N., Demers, E. G., Crocker, A. W. & Hogan, D. A. Use of RNA–protein complexes for genome editing in non-albicans Candida species. mSphere 2, e00218-17 (2017).
Liu, Q. et al. Development of a genome-editing CRISPR/Cas9 system in thermophilic fungal Myceliophthora species and its application to hyper-cellulase production strain engineering. Biotechnol. Biofuels 10, 1 (2017).
Palermo, G., Miao, Y., Walker, R. C., Jinek, M. & McCammon, J. A. CRISPR-Cas9 conformational activation as elucidated from enhanced molecular simulations. Proc. Natl Acad. Sci. USA 114, 7260–7265 (2017).
Ivics, Z. & Izsvak, Z. Sleeping Beauty transposition. Microbiol. Spectr. 3, MDNA3-0042-2014 (2015).
Wang, W. et al. Chromosomal transposition of PiggyBac in mouse embryonic stem cells. Proc. Natl Acad. Sci. USA 105, 9290–9295 (2008).
Adey, A. et al. Rapid, low-input, low-bias construction of shotgun fragment libraries by high-density in vitro transposition. Genome Biol. 11, R119 (2010).
Potapov, V. & Ong, J. L. Examining sources of error in PCR by single-molecule sequencing. PLoS ONE 12, e0169774 (2017).
Aird, D. et al. Analyzing and minimizing PCR amplification bias in Illumina sequencing libraries. Genome Biol. 12, R18 (2011).
Acknowledgements
This work was supported by the Thousand Young Talents Program (J.W.), Ministry of Science and Technology of China (2016YFC0900103 to J.W.), National Natural Science Foundation of China (21675098 to J.W.), THU-PKU Center for Life Sciences (J.G. and J.W.) and the Agency for Sciences, Technology and Research of Singapore (NMRC/OFIRG/0072/2018 to Y.W.). J.G. thanks T. Ha for her continued support and company.
Author information
Authors and Affiliations
Contributions
J.G. developed and wrote the protocol; H.W. and Z.L. performed computational analyses; C.C. tested the reproducibility of the protocol; A.H.-H.W. provided technical assistance; and J.G., J.W., and Y.W. conceptualized, designed and supervised the study. All authors contributed to editing the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Protocols thanks Damian Krysan and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key reference using this protocol
Gao, J. et al. Nat. Commun. 9, 4495 (2018): https://www.nature.com/articles/s41467-018-06944-1
Supplementary information
Supplementary Information
Supplementary Results, Supplementary Fig. 1 and Supplementary Tables 1 and 2.
Supplementary Data 1
The haploid C. albicans GZY892 assembly sequence and its annotation file.
Supplementary Data 2
Output file contains the processed mapping results from Step 69.
Supplementary Data 3
Output file lists genes mapped by insertions from Step 69.
Rights and permissions
About this article
Cite this article
Li, Z., Wang, H., Cai, C. et al. Genome-wide piggyBac transposon-based mutagenesis and quantitative insertion-site analysis in haploid Candida species. Nat Protoc 15, 2705–2727 (2020). https://doi.org/10.1038/s41596-020-0351-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-020-0351-3
This article is cited by
-
LncRNA DINOR is a virulence factor and global regulator of stress responses in Candida auris
Nature Microbiology (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.