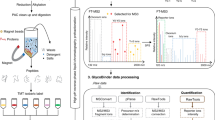
Abstract
The glycocalyx comprises glycosylated proteins and lipids and fcorms the outermost layer of cells. It is involved in fundamental inter- and intracellular processes, including non-self-cell and self-cell recognition, cell signaling, cellular structure maintenance, and immune protection. Characterization of the glycocalyx is thus essential to understanding cell physiology and elucidating its role in promoting health and disease. This protocol describes how to comprehensively characterize the glycocalyx N-glycans and O-glycans of glycoproteins, as well as intact glycolipids in parallel, using the same enriched membrane fraction. Profiling of the glycans and the glycolipids is performed using nanoflow liquid chromatography–mass spectrometry (nanoLC-MS). Sample preparation, quantitative LC–tandem MS (LC-MS/MS) analysis, and data processing methods are provided. In addition, we discuss glycoproteomic analysis that yields the site-specific glycosylation of membrane proteins. To reduce the amount of sample needed, N-glycan, O-glycan, and glycolipid analyses are performed on the same enriched fraction, whereas glycoproteomic analysis is performed on a separate enriched fraction. The sample preparation process takes 2–3 d, whereas the time spent on instrumental and data analyses could vary from 1 to 5 d for different sample sizes. This workflow is applicable to both cell and tissue samples. Systematic changes in the glycocalyx associated with specific glycoforms and glycoconjugates can be monitored with quantitation using this protocol. The ability to quantitate individual glycoforms and glycoconjugates will find utility in a broad range of fundamental and applied clinical studies, including glycan-based biomarker discovery and therapeutics.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout









Similar content being viewed by others
Data availability
The data are all available online. Data from ref. 23 (Park, D. et al. Glycobiology 27, 847–860 (2017)) (used for Fig. 4a) are available at https://doi.org/10.1093/glycob/cwx041. Data from ref. 24 (Park, D. et al. Chem. Sci. 9, 6271–6285 (2018)) (used for Figs. 4b and 9) are available at https://doi.org/10.1039/c8sc01875h. Data from ref. 26 (Wong, M. et al. Sci. Rep. 8, 10993 (2018)) (used for Fig. 4c) are available at https://doi.org/10.1038/s41598-018-29324-7. Data from ref. 25 (Li, Q. et al. Chem. Sci. 10, 6199–6209 (2019)) (used for Fig. 6) are available at https://doi.org/10.1039/c9sc01360a. Data from ref. 33 (Park, D. et al. Mol. Cell. Proteomics 14, 2910–2921 (2015)) (used for Fig. 8) are available at https://doi.org/10.1074/mcp.M115.053983.
References
Martinez-Seara Monne, H., Danne, R., Róg, T., Ilpo, V. & Gurtovenko, A. Structure of glycocalyx. Biophys. J. 104, 251a (2013).
Reitsma, S., Slaaf, D. W., Vink, H., Van Zandvoort, M. A. & Oude Egbrink, M. G. The endothelial glycocalyx: composition, functions, and visualization. Pflügers Arch. 454, 345–359 (2007).
Flessner, M. F. Endothelial glycocalyx and the peritoneal barrier. Perit. Dial. Int. 28, 6–12 (2008).
Mensah, S. A. et al. Regeneration of glycocalyx by heparan sulfate and sphingosine 1-phosphate restores inter-endothelial communication. PloS ONE 12, e0186116 (2017).
Kuo, J. C.-H., Gandhi, J. G., Zia, R. N. & Paszek, M. J. Physical biology of the cancer cell glycocalyx. Nat. Phys. 14, 658 (2018).
Yao, Y., Rabodzey, A. & Dewey, C. F. Jr Glycocalyx modulates the motility and proliferative response of vascular endothelium to fluid shear stress. Am. J. Physiol. Heart Circ. Physiol. 293, H1023–H1030 (2007).
Gristina, A. G. & Costerton, J. Bacterial adherence and the glycocalyx and their role in musculoskeletal infection. Orthop. Clin. North Am. 15, 517–535 (1984).
Martin, L., Koczera, P., Zechendorf, E. & Schuerholz, T. The endothelial glycocalyx: new diagnostic and therapeutic approaches in sepsis. BioMed. Res. Int. 2016, 1–8 (2016).
Arabyan, N. et al. Salmonella degrades the host glycocalyx leading to altered infection and glycan remodeling. Sci. Rep. 6, 29525 (2016).
Yeo, T. W. et al. Glycocalyx breakdown is associated with severe disease and fatal outcome in Plasmodium falciparum malaria. Clin. Infect. Dis. 69, 1712–1720 (2019).
Hakomori, S.-i. in Advances in Cancer Research Vol. 52 (eds Vande Woude, G. F. & Klein, G.) 257–331 (Academic Press, 1989).
Noda, K. et al. Relationship between elevated FX expression and increased production of GDP-l-fucose, a common donor substrate for fucosylation in human hepatocellular carcinoma and hepatoma cell lines. Cancer Res. 63, 6282–6289 (2003).
Nie, H. et al. Specific N-glycans of hepatocellular carcinoma cell surface and the abnormal increase of core-α-1, 6-fucosylated triantennary glycan via N-acetylglucosaminyltransferases-IVa regulation. Sci. Rep. 5, 16007 (2015).
Paszek, M. J. et al. The cancer glycocalyx mechanically primes integrin-mediated growth and survival. Nature 511, 319 (2014).
Maverakis, E. et al. Glycans in the immune system and The Altered Glycan Theory of Autoimmunity: a critical review. J. Autoimmun. 57, 1–13 (2015).
Dixon, J. et al. Electron microscopic investigation of the bladder urothelium and glycocalyx in patients with interstitial cystitis. J. Urol. 135, 621–625 (1986).
de Buy Wenniger, L. J. M. et al. The cholangiocyte glycocalyx stabilizes the ‘biliary HCO3-umbrella’: an integrated line of defense against toxic bile acids. Dig. Dis. 33, 397–407 (2015).
An, H. J., Kronewitter, S. R., de Leoz, M. L. A. & Lebrilla, C. B. Glycomics and disease markers. Curr. Opin. Chem. Biol. 13, 601–607 (2009).
Blomme, B., Van Steenkiste, C., Callewaert, N. & Van Vlierberghe, H. Alteration of protein glycosylation in liver diseases. J. Hepatol. 50, 592–603 (2009).
Dall’Olio, F. et al. N-glycomic biomarkers of biological aging and longevity: a link with inflammaging. Ageing Res. Rev. 12, 685–698 (2013).
Lee, S.-M. et al. N-Glycosylation of asparagine 130 in the extracellular domain of the human calcitonin receptor significantly increases peptide hormone affinity. Biochemistry 56, 3380–3393 (2017).
Veillon, L., Fakih, C., Abou-El-Hassan, H., Kobeissy, F. & Mechref, Y. Glycosylation changes in brain cancer. ACS Chem. Neurosci. 9, 51–72 (2017).
Park, D. et al. Enterocyte glycosylation is responsive to changes in extracellular conditions: implications for membrane functions. Glycobiology 27, 847–860 (2017).
Park, D. D. et al. Membrane glycomics reveal heterogeneity and quantitative distribution of cell surface sialylation. Chem. Sci. 9, 6271–6285 (2018).
Li, Q., Xie, Y., Xu, G. & Lebrilla, C. B. Identification of potential sialic acid binding proteins on cell membranes by proximity chemical labeling. Chem. Sci. 10, 6199–6209 (2019).
Wong, M., Xu, G., Park, D., Barboza, M. & Lebrilla, C. B. Intact glycosphingolipidomic analysis of the cell membrane during differentiation yields extensive glycan and lipid changes. Sci. Rep. 8, 10993 (2018).
Wu, S., Tao, N., German, J. B., Grimm, R. & Lebrilla, C. B. Development of an annotated library of neutral human milk oligosaccharides. J. Proteome Res. 9, 4138–4151 (2010).
Ninonuevo, M. R. et al. A strategy for annotating the human milk glycome. J. Agric. Food Chem. 54, 7471–7480 (2006).
Barboza, M. et al. Glycosylation of human milk lactoferrin exhibits dynamic changes during early lactation enhancing its role in pathogenic bacteria-host interactions. Mol. Cell. Proteom. 11, M111. 015248 (2012).
Chu, C. S. et al. Profile of native N‐linked glycan structures from human serum using high performance liquid chromatography on a microfluidic chip and time‐of‐flight mass spectrometry. Proteomics 9, 1939–1951 (2009).
Lee, H. et al. Multiple precursor ion scanning of gangliosides and sulfatides with a reversed-phase microfluidic chip and quadrupole time-of-flight mass spectrometry. Anal. Chem. 84, 5905–5912 (2012).
An, H. J. et al. Extensive determination of glycan heterogeneity reveals an unusual abundance of high mannose glycans in enriched plasma membranes of human embryonic stem cells. Mol. Cell. Proteom. 11, M111. 010660 (2012).
Park, D. et al. Characteristic changes in cell surface glycosylation accompany intestinal epithelial cell (IEC) differentiation: high mannose structures dominate the cell surface glycome of undifferentiated enterocytes. Mol. Cell. Proteom. 14, 2910–2921 (2015).
Ruhaak, L. R. et al. Differential N-glycosylation patterns in lung adenocarcinoma tissue. J. Proteome Res. 14, 4538–4549 (2015).
Ruhaak, L. et al. Glycan labeling strategies and their use in identification and quantification. Anal. Bioanal. l Chem. 397, 3457–3481 (2010).
Goldstein, I. J., Winter, H. C. & Poretz, R. D. in New Comprehensive Biochemistry Vol. 29 (eds Montreuil, J., Vliegenthart, J. F. G. & Schachter, H.) 403–474 (Elsevier, 1997).
Lee, L. Y. et al. An optimized approach for enrichment of glycoproteins from cell culture lysates using native multi‐lectin affinity chromatography. J. Sep. Sci. 35, 2445–2452 (2012).
Palaniappan, K. K. & Bertozzi, C. R. Chemical glycoproteomics. Chem. Rev. 116, 14277–14306 (2016).
Kuhn, P. H. et al. Secretome protein enrichment identifies physiological BACE1 protease substrates in neurons. EMBO J. 31, 3157–3168 (2012).
Haun, R. S. et al. Bioorthogonal labeling cell-surface proteins expressed in pancreatic cancer cells to identify potential diagnostic/therapeutic biomarkers. Cancer Biol. Ther. 16, 1557–1565 (2015).
Zacharias, L. G. et al. HILIC and ERLIC enrichment of glycopeptides derived from breast and brain cancer cells. J. Proteome Res. 15, 3624–3634 (2016).
Sibille, E. et al. Ganglioside profiling of the human retina: comparison with other ocular structures, brain and plasma reveals tissue specificities. PloS ONE 11, e0168794 (2016).
Lapainis, T., Rubakhin, S. S. & Sweedler, J. V. Capillary electrophoresis with electrospray ionization mass spectrometric detection for single-cell metabolomics. Anal. Chem. 81, 5858–5864 (2009).
Mellors, J., Gorbounov, V., Ramsey, R. & Ramsey, J. Fully integrated glass microfluidic device for performing high-efficiency capillary electrophoresis and electrospray ionization mass spectrometry. Anal. Chem. 80, 6881–6887 (2008).
Ruhaak, L. R., Xu, G., Li, Q., Goonatilleke, E. & Lebrilla, C. B. Mass spectrometry approaches to glycomic and glycoproteomic analyses. Chem. Rev. 118, 7886–7930 (2018).
Both, P. et al. Discrimination of epimeric glycans and glycopeptides using IM-MS and its potential for carbohydrate sequencing. Nat. Chem. 6, 65 (2014).
Plasencia, M. D., Isailovic, D., Merenbloom, S. I., Mechref, Y. & Clemmer, D. E. Resolving and assigning N-linked glycan structural isomers from ovalbumin by IMS-MS. J. Am. Soc. Mass Spectrom. 19, 1706–1715 (2008).
Hofmann, J. et al. Identification of Lewis and blood group carbohydrate epitopes by ion mobility-tandem-mass spectrometry fingerprinting. Anal. Chem. 89, 2318–2325 (2017).
Leopold, J., Popkova, Y., Engel, K. M. & Schiller, J. Recent developments of useful MALDI matrices for the mass spectrometric characterization of lipids. Biomolecules 8, 173 (2018).
Seipert, R. R. et al. Factors that influence fragmentation behavior of N-linked glycopeptide ions. Anal. Chem. 80, 3684–3692 (2008).
Cao, L. et al. Intact glycopeptide characterization using mass spectrometry. Expert Rev. Proteom. 13, 513–522 (2016).
Yu, Q. et al. Electron-transfer/higher-energy collision dissociation (EThcD)-enabled intact glycopeptide/glycoproteome characterization. J. Am. Soc. Mass Spectrom. 28, 1751–1764 (2017).
Forgue-Lafitte, M.-E., Coudray, A.-M., Bréant, B. & Mešter, J. Proliferation of the human colon carcinoma cell line HT29: autocrine growth and deregulated expression of the c-myc oncogene. Cancer Res. 49, 6566–6571 (1989).
Dey, P. M., Brownleader, M. D. & Harborne, J. B. in Plant Biochemistry (eds Dey, P. M. & Harborne, J. B.) 1–47 (Academic Press, 1997).
Banneau, G., Ayadi, M., Armenoult, L. & Carvalho, E. Homogenization of cartilage tumors to extract total RNA to microarray and sequencing analysis using Precellys bead-beating technology. Biotechniques 52, 196–197 (2012).
Fujiki, Y., Hubbard, A. L., Fowler, S. & Lazarow, P. B. Isolation of intracellular membranes by means of sodium carbonate treatment: application to endoplasmic reticulum. J. Cell Biol. 93, 97–102 (1982).
Li, Q., Xie, Y., Wong, M. & Lebrilla, B. C. Characterization of cell glycocalyx with mass spectrometry methods. Cells 8, 882 (2019).
Sandoval, W. N. et al. Rapid removal of N-linked oligosaccharides using microwave assisted enzyme catalyzed deglycosylation. Int. J. Mass Spectrom. 259, 117–123 (2007).
Lauber, M. A. et al. Rapid preparation of released N-glycans for HILIC analysis using a labeling reagent that facilitates sensitive fluorescence and ESI-MS detection. Anal. Chem. 87, 5401–5409 (2015).
Van Ree, R. et al. β(1, 2)-xylose and α(1, 3)-fucose residues have a strong contribution in IgE binding to plant glycoallergens. J. Biol. Chem. 275, 11451–11458 (2000).
Hua, S. et al. Comprehensive native glycan profiling with isomer separation and quantitation for the discovery of cancer biomarkers. Analyst 136, 3663–3671 (2011).
Park, D. et al. Salmonella typhimurium enzymatically landscapes the host intestinal epithelial cell (IEC) surface glycome to increase invasion. Mol. Cell. Proteom. 15, 3653–3664 (2016).
Folch, J., Lees, M. & Sloane-Stanley, G. A simple method for the isolation and purification of total lipids from animal tissues. J. Biol. Chem. 226, 497–509 (1957).
Bligh, E. G. & Dyer, W. J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37, 911–917 (1959).
Miura, Y. et al. Glycoblotting-assisted O-glycomics: ammonium carbamate allows for highly efficient O-glycan release from glycoproteins. Anal. Chem. 82, 10021–10029 (2010).
Sun, S., Zhou, J.-Y., Yang, W. & Zhang, H. Inhibition of protein carbamylation in urea solution using ammonium-containing buffers. Anal. Biochem. 446, 76–81 (2014).
Li, Q. et al. Site-specific glycosylation quantitation of 50 serum glycoproteins enhanced by predictive glycopeptidomics for improved disease biomarker discovery. Anal. Chem. 91, 5433–5445 (2019).
Du, Y., Wang, F., May, K., Xu, W. & Liu, H. LC–MS analysis of glycopeptides of recombinant monoclonal antibodies by a rapid digestion procedure. J. Chromatogr. B 907, 87–93 (2012).
Kalli, A., Smith, G. T., Sweredoski, M. J. & Hess, S. Evaluation and optimization of mass spectrometric settings during data-dependent acquisition mode: focus on LTQ-Orbitrap mass analyzers. J. Proteome Res. 12, 3071–3086 (2013).
Kalli, A. & Hess, S. Effect of mass spectrometric parameters on peptide and protein identification rates for shotgun proteomic experiments on an LTQ‐Orbitrap mass analyzer. Proteomics 12, 21–31 (2012).
Yang, H., Yang, C. & Sun, T. Characterization of glycopeptides using a stepped higher‐energy C‐trap dissociation approach on a hybrid quadrupole orbitrap. Rapid Commun. Mass Spectrom. 32, 1353–1362 (2018).
Chen, Z. et al. Site-specific characterization and quantitation of N-glycopeptides in PKM2 knockout breast cancer cells using DiLeu isobaric tags enabled by electron-transfer/higher-energy collision dissociation (EThcD). Analyst 143, 2508–2519 (2018).
Singh, C., Zampronio, C. G., Creese, A. J. & Cooper, H. J. Higher energy collision dissociation (HCD) product ion-triggered electron transfer dissociation (ETD) mass spectrometry for the analysis of N-linked glycoproteins. J. Proteome Res. 11, 4517–4525 (2012).
Riley, N. M., Hebert, A. S., Westphall, M. S. & Coon, J. J. Capturing site-specific heterogeneity with large-scale N-glycoproteome analysis. Nat. Commun. 10, 1311 (2019).
Kronewitter, S. R. et al. The development of retrosynthetic glycan libraries to profile and classify the human serum N-linked glycome. Proteomics 9, 2986–2994 (2009).
Senger, R. S. & Karim, M. N. Prediction of N-linked glycan branching patterns using artificial neural networks. Math. Biosci. 211, 89–104 (2008).
Jackson, S. & Nicolson, S. W. Xylose as a nectar sugar: from biochemistry to ecology. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 131, 613–620 (2002).
Samraj, A. N. et al. A red meat-derived glycan promotes inflammation and cancer progression. Proc. Natl Acad. Sci. USA 112, 542–547 (2015).
Chou, H.-H. et al. A mutation in human CMP-sialic acid hydroxylase occurred after the Homo-Pan divergence. Proc. Natl Acad. Sci. USA 95, 11751–11756 (1998).
Tra, V. N. & Dube, D. H. Glycans in pathogenic bacteria–potential for targeted covalent therapeutics and imaging agents. Chem. Commun. 50, 4659–4673 (2014).
Yamakawa, N. et al. Systems glycomics of adult zebrafish identifies organ-specific sialylation and glycosylation patterns. Nat. Commun. 9, 4647 (2018).
Hartley, M. D. et al. Biochemical characterization of the O-linked glycosylation pathway in Neisseria gonorrhoeae responsible for biosynthesis of protein glycans containing N, N′-diacetylbacillosamine. Biochemistry 50, 4936–4948 (2011).
Merrill, A. H. SphinGOMAP: Lipidomic analysis of [glyco]sphingolipid metabolism. FASEB J. 20, A1472 (2006).
Zeng, W.-F. et al. pGlyco: a pipeline for the identification of intact N-glycopeptides by using HCD-and CID-MS/MS and MS3. Sci. Rep. 6, 25102 (2016).
Bern, M., Kil, Y. J. & Becker, C. in Current Protocols in Bioinformatics (Wiley, 2002).
Aldredge, D., An, H. J., Tang, N., Waddell, K. & Lebrilla, C. B. Annotation of a serum N-glycan library for rapid identification of structures. J. Proteome Res. 11, 1958–1968 (2012).
Song, T., Aldredge, D. & Lebrilla, C. B. A method for in-depth structural annotation of human serum glycans that yields biological variations. Anal. Chem. 87, 7754–7762 (2015).
Hülsmeier, A. J., Paesold-Burda, P. & Hennet, T. N-glycosylation site occupancy in serum glycoproteins using multiple reaction monitoring liquid chromatography-mass spectrometry. Mol. Cell. Proteom. 6, 2132–2138 (2007).
Zhang, F., Zhang, Z. & Linhardt, R. J. in Handbook of Glycomics (eds Cummings, R. D. & Pierce, J. M.) 59–80 (Academic Press, 2010).
Xu, G. et al. Unveiling the metabolic fate of monosaccharides in cell membranes with glycomic and glycoproteomic analyses. Chem. Sci. 10, 6992–7002 (2019).
Acknowledgements
This work was supported by the National Institutes of Health (GMRO1R01, GM049077).
Author information
Authors and Affiliations
Contributions
Q.L., Y.X., M.W., M.B., and C.B.L. contributed to the development of this protocol and wrote and edited the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key references using this protocol
Park, D. D. et al. Chem. Sci. 9, 6271–6285 (2018): https://doi.org/10.1039/c8sc01875h
Wong, M., Xu, G., Park, D., Barboza, M. & Lebrilla, C. B. Sci. Rep. 8, 10993 (2018): https://doi.org/10.1038/s41598-018-29324-7
Li, Q., Xie, Y., Xu, G. & Lebrilla, C. B. Chem. Sci. 10, 6199–6209 (2019): https://doi.org/10.1039/c9sc01360a
Supplementary information
Supplementary Information
Supplementary Figs 1 and 2.
Supplementary Data 1
The table contains examples of N-glycans with their masses, compositions, and types.
Supplementary Data 2
The table contains possible ceramide m/z values.
Supplementary Data 3
The table can be used to match the closest glycan composition to the glycosphingolipid with a known precursor m/z, precursor charge state, and ceramide m/z.
Supplementary Data 4
The table contains some common saccharide and lipid fragments.
Supplementary Data 5
The table contains a pivot table for generating a database of glycosphingolipids.
Rights and permissions
About this article
Cite this article
Li, Q., Xie, Y., Wong, M. et al. Comprehensive structural glycomic characterization of the glycocalyxes of cells and tissues. Nat Protoc 15, 2668–2704 (2020). https://doi.org/10.1038/s41596-020-0350-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-020-0350-4
This article is cited by
-
Mouse tissue glycome atlas 2022 highlights inter-organ variation in major N-glycan profiles
Scientific Reports (2022)
-
On-tissue amidation of sialic acid with aniline for sensitive imaging of sialylated N-glycans from FFPE tissue sections via MALDI mass spectrometry
Analytical and Bioanalytical Chemistry (2022)
-
Toward robust N-glycomics of various tissue samples that may contain glycans with unknown or unexpected structures
Scientific Reports (2021)
-
Importance of evaluating protein glycosylation in pluripotent stem cell-derived cardiomyocytes for research and clinical applications
Pflügers Archiv - European Journal of Physiology (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.