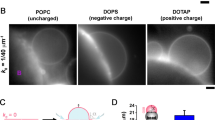
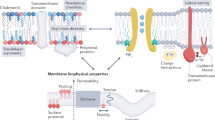
Abstract
Cellular membrane processes, from signal transduction to membrane fusion and fission, depend on acute membrane deformations produced by small and short-lived protein complexes working in conditions far from equilibrium. Real-time monitoring and quantitative assessment of such deformations are challenging; hence, mechanistic analyses of the protein action are commonly based on ensemble averaging, which masks important mechanistic details of the action. In this protocol, we describe how to reconstruct and quantify membrane remodeling by individual proteins and small protein complexes in vitro, using an ultra-short (80- to 400-nm) lipid nanotube (usNT) template. We use the luminal conductance of the usNT as the real-time reporter of the protein interaction(s) with the usNT. We explain how to make and calibrate the usNT template to achieve subnanometer precision in the geometrical assessment of the molecular footprints on the nanotube membrane. We next demonstrate how membrane deformations driven by purified proteins implicated in cellular membrane remodeling can be analyzed at a single-molecule level. The preparation of one usNT takes ~1 h, and the shortest procedure yielding the basic geometrical parameters of a small protein complex takes 10 h.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout










Similar content being viewed by others
References
Groves, J. T. Bending mechanics and molecular organization in biological membranes. Annu. Rev. Phys. Chem. 58, 697–717 (2007).
Bassereau, P. et al. The 2018 biomembrane curvature and remodeling roadmap. J. Phys. D. Appl. Phys. 51, 343001 (2018).
Roux, A. et al. Membrane curvature controls dynamin polymerization. Proc. Natl Acad. Sci. USA 107, 4141–4146 (2010).
Pinot, M., Goud, B. & Manneville, J.-B. Physical aspects of COPI vesicle formation. Mol. Membr. Biol. 27, 428–442 (2010).
Capraro, B. R. et al. Kinetics of endophilin N-BAR domain dimerization and membrane interactions. J. Biol. Chem. 288, 12533–12543 (2013).
Shnyrova, A. V., Frolov, V. A. & Zimmerberg, J. Domain-driven morphogenesis of cellular membranes. Curr. Biol. 19, R772–R780 (2009).
Groves, J. T. & Kuriyan, J. Molecular mechanisms in signal transduction at the membrane. Nat. Struct. Mol. Biol. 17, 659–665 (2010).
Zimmerberg, J. & Kozlov, M. M. How proteins produce cellular membrane curvature. Nat. Rev. Mol. Cell Biol. 7, 9–19 (2006).
McMahon, H. T. & Gallop, J. L. Membrane curvature and mechanisms of dynamic cell membrane remodelling. Nature 438, 590–596 (2005).
Zhu, C., Das, S. L. & Baumgart, T. Nonlinear sorting, curvature generation, and crowding of endophilin N-BAR on tubular membranes. Biophys. J. 102, 1837–1845 (2012).
Callan-Jones, A., Sorre, B. & Bassereau, P. Curvature-driven lipid sorting in biomembranes. Cold Spring Harb. Perspect. Biol. 3, 1–14 (2011).
Hsieh, W. T. et al. Curvature sorting of peripheral proteins on solid-supported wavy membranes. Langmuir 28, 12838–12843 (2012).
Antonny, B. et al. Membrane fission by dynamin: what we know and what we need to know. EMBO J. 35, 2270–2284 (2016).
Jia, H. & Schwille, P. Bottom-up synthetic biology: reconstitution in space and time. Curr. Opin. Biotechnol. 60, 179–187 (2019).
Liu, A. P. & Fletcher, D. A. Biology under construction: in vitro reconstitution of cellular function. Nat. Rev. Mol. Cell Biol. 10, 644–650 (2009).
Huang, S.-C. J. et al. Formation, stability, and mobility of one-dimensional lipid bilayers on polysilicon nanowires. Nano Lett. 7, 3355–3359 (2007).
Zhou, X., Moran-Mirabal, J. M., Craighead, H. G. & McEuen, P. L. Supported lipid bilayer/carbon nanotube hybrids. Nat. Nanotechnol. 2, 185–190 (2007).
Artyukhin, A. B. et al. Functional one-dimensional lipid bilayers on carbon nanotube templates. J. Am. Chem. Soc. 127, 7538–7542 (2005).
Li, X. et al. A nanostructure platform for live-cell manipulation of membrane curvature. Nat. Protoc. 14, 1772–1802 (2019).
Shi, Z. & Baumgart, T. Dynamics and instabilities of lipid bilayer membrane shapes. Adv. Colloid Interface Sci. 208, 76–88 (2014).
Shnyrova, A. V. et al. Geometric catalysis of membrane fission driven by flexible dynamin rings. Science 339, 1433–1436 (2013).
Harmandaris, V. A. & Deserno, M. A novel method for measuring the bending rigidity of model lipid membranes by simulating tethers. J. Chem. Phys. 125, 204905 (2006).
Fang, Y., Rand, R. P., Leikin, S. & Kozlov, M. M. Chain-melting reentrant transition in bimolecular layers at large separations. Phys. Rev. Lett. 70, 3623–3626 (1993).
Frolov, V. A., Lizunov, V. A., Dunina-Barkovskaya, A. Y., Samsonov, A. V. & Zimmerberg, J. Shape bistability of a membrane neck: a toggle switch to control vesicle content release. Proc. Natl Acad. Sci. USA 100, 8698–8703 (2003).
Houghtaling, J. et al. Estimation of shape, volume, and dipole moment of individual proteins freely transiting a synthetic nanopore. ACS Nano 13, 5231–5242 (2019).
Bashkirov, P. V. et al. GTPase cycle of dynamin is coupled to membrane squeeze and release, leading to spontaneous fission. Cell 135, 1276–1286 (2008).
Roux, A. The physics of membrane tubes: soft templates for studying cellular membranes. Soft Matter 9, 6726–6736 (2013).
Simunovic, M., Prévost, C., Andrew, C. J. & Bassereau, P. Physical basis of some membrane shaping mechanisms. Philos. Trans. R. Soc. Lond. A 374, 20160034 (2016).
Rosenboom, H. & Lindau, M. Exo-endocytosis and closing of the fission pore during endocytosis in single pituitary nerve terminals internally perfused with high calcium concentrations. Proc. Natl Acad. Sci. 91, 5267–5271 (1994).
Debus, K. & Lindau, M. Resolution of patch capacitance recordings and of fusion pore conductances in small vesicles. Biophys. J. 78, 2983–2997 (2000).
Velasco-Olmo, A., Ormaetxea Gisasola, J., Martinez Galvez, J. M., Vera Lillo, J. & Shnyrova, A. V. Combining patch-clamping and fluorescence microscopy for quantitative reconstitution of cellular membrane processes with giant suspended bilayers. Sci. Rep. 9, 7255 (2019).
Bashkirov, P. V., Chekashkina, K. V., Akimov, S. A., Kuzmin, P. I. & Frolov, V. A. Variation of lipid membrane composition caused by strong bending. Biochem. Mosc. Suppl. Ser. A 5, 205–211 (2011).
Bashkirov, P. V. Membrane nanotubes in the electric field as a model for measurement of mechanical parameters of the lipid bilayer. Biochem. Mosc. Suppl. Ser. A 1, 176–184 (2007).
Chappie, J. S. et al. A pseudoatomic model of the dynamin polymer identifies a hydrolysis-dependent powerstroke. Cell 147, 209–222 (2011).
Stowell, M. H. B., Marks, B., Wigge, P. & McMahon, H. T. Nucleotide-dependent conformational changes in dynamin: evidence for a mechanochemical molecular spring. Nat. Cell Biol. 1, 27–32 (1999).
Ando, T. High-speed atomic force microscopy and its future prospects. Biophys. Rev. 10, 285–292 (2018).
Colom, A., Redondo-Morata, L., Chiaruttini, N., Roux, A. & Scheuring, S. Dynamic remodeling of the dynamin helix during membrane constriction. Proc. Natl Acad. Sci. USA 114, 5449–5454 (2017).
Takeda, T. et al. Dynamic clustering of dynamin-amphiphysin helices regulates membrane constriction and fission coupled with GTP hydrolysis. Elife 7, e30246 (2018).
Vicidomini, G., Bianchini, P. & Diaspro, A. STED super-resolved microscopy. Nat. Methods 15, 173–182 (2018).
Dar, S., Kamerkar, S. C. & Pucadyil, T. J. A high-throughput platform for real-time analysis of membrane fission reactions reveals the mechanism of dynamin function. Nat. Cell Biol. 17, 1588–1596 (2015).
Madsen, K. L., Bhatia, V. K., Gether, U. & Stamou, D. BAR domains, amphipathic helices and membrane-anchored proteins use the same mechanism to sense membrane curvature. FEBS Lett. 584, 1848–1855 (2010).
Espadas, J. et al. Dynamic constriction and fission of endoplasmic reticulum membranes by reticulon. Nat. Commun. 10, 5327 (2019).
Simunovic, M. et al. Friction nediates scission of tubular membranes scaffolded by BAR proteins. Cell 170, 172–184.e11 (2017).
Schmid, S. L. & Frolov, V. A. Dynamin: functional design of a membrane fission catalyst. Annu. Rev. Cell Dev. Biol. 27, 79–105 (2011).
Ferguson, S. M. & De Camilli, P. Dynamin, a membrane-remodelling GTPase. Nat. Rev. Mol. Cell Biol. 13, 75–88 (2012).
Mattila, J. P. et al. A hemi-fission intermediate links two mechanistically distinct stages of membrane fission. Nature 524, 109–113 (2015).
Capraro, B. R., Yoon, Y., Cho, W. & Baumgart, T. Curvature sensing by the epsin N-terminal homology domain measured on cylindrical lipid membrane tethers. J. Am. Chem. Soc. 132, 1200–1201 (2010).
Tunuguntla, R. H., Escalada, A., A Frolov, V. & Noy, A. Synthesis, lipid membrane incorporation, and ion permeability testing of carbon nanotube porins. Nat. Protoc. 11, 2029–2047 (2016).
Kong, L. et al. Cryo-EM of the dynamin polymer assembled on lipid membrane. Nature 560, 258–262 (2018).
Gracià, R. S., Bezlyepkina, N., Knorr, R. L., Lipowsky, R. & Dimova, R. Effect of cholesterol on the rigidity of saturated and unsaturated membranes: fluctuation and electrodeformation analysis of giant vesicles. Soft Matter 6, 1472–1482 (2010).
Pan, J., Mills, T. T., Tristram-Nagle, S. & Nagle, J. F. Cholesterol perturbs lipid bilayers nonuniversally. Phys. Rev. Lett. 100, 198103 (2008).
Acknowledgements
This work was partially supported by NIH R01GM121725 to V.A.F.; Spanish Ministry of Science, Innovation and Universities and FEDER grant BFU2015-70552-P to V.A.F. and A.V.S.; Russian Science Foundation grant 17-75-30064 to P.V.B.; and the Fundación Biofísica Bizkaia and the Basque Excellence Research Centre (BERC) program of the Basque Government and the Ministry of Science and Higher Education of the Russian Federation.
Author information
Authors and Affiliations
Contributions
Conceptualization, V.A.F., P.V.B., A.V.S.; methodology, V.A.F., P.V.B.; investigation, K.C., P.A., J.V.L., P.V.B.; theoretical analysis, V.A.F., P.I.K., P.V.B.; writing of original draft, V.A.F., P.V.B., A.V.S.; review and editing, all authors; funding acquisition, V.A.F., P.V.B., A.V.S.; resources, V.A.F., P.V.B., A.V.S.; supervision, V.A.F., P.V.B., A.V.S.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Protocols thanks Patricia Bassereau and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key references using this protocol
Frolov, V. A. et al. Proc. Natl Acad. Sci. USA 100, 8698–8703 (2003): https://www.pnas.org/content/100/15/8698
Bashkirov, P. V. et al. Cell 135, 1276–1286 (2008): https://www.cell.com/cell/fulltext/S0092-8674(08)01503-1
Shnyrova, A. V. et al. Science. 339, 1433–1436 (2013): https://science.sciencemag.org/content/339/6126/1433.full
Mattila, J.-P. et al. Nature 524, 109–113 (2015): https://www.nature.com/articles/nature14509
Supplementary information
Source data
Source Data Fig. 5
Statistical source data.
Source Data Fig. 7
Source data (two columns x and y axes).
Source Data Fig. 8
Source data (two columns x and y axes).
Source Data Fig. 9
Statistical source data (two columns for gray and black histograms).
Source Data Fig. 10
Statistical source data (for the histograms in Fig. 10a and b).
Rights and permissions
About this article
Cite this article
Bashkirov, P.V., Kuzmin, P.I., Chekashkina, K. et al. Reconstitution and real-time quantification of membrane remodeling by single proteins and protein complexes. Nat Protoc 15, 2443–2469 (2020). https://doi.org/10.1038/s41596-020-0337-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-020-0337-1
This article is cited by
-
Membrane activity of viral proteins
Biophysical Reviews (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.