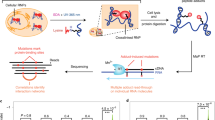
Abstract
Long noncoding RNAs (lncRNAs) are recently discovered transcripts that regulate vital cellular processes, such as cellular differentiation and DNA replication, and are crucially connected to diseases. Although the 3D structures of lncRNAs are key determinants of their function, the unprecedented molecular complexity of lncRNAs has so far precluded their 3D structural characterization at high resolution. It is thus paramount to develop novel approaches for biochemical and biophysical characterization of these challenging targets. Here, we present a protocol that integrates non-denaturing lncRNA purification with in-solution hydrodynamic analysis and single-particle atomic force microscopy (AFM) imaging to produce highly homogeneous lncRNA preparations and visualize their 3D topology at ~15-Å resolution. Our protocol is suitable for imaging lncRNAs in biologically active conformations and for measuring structural defects of functionally inactive mutants that have been identified by cell-based functional assays. Once optimized for the specific target lncRNA of choice, our protocol leads from cloning to AFM imaging within 3–4 weeks and can be implemented using state-of-the-art biochemical and biophysical instrumentation by trained researchers familiar with RNA handling and supported by AFM and small-angle X-ray scattering (SAXS) experts.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
All data generated or analyzed during this study are included in the paper and its Supplementary Information and are available from the corresponding author on request. We have deposited in figshare an extensive set of AFM images, which we have used for calculation of the PSDs:
MEG3 v1 compact form: https://figshare.com/s/e73aff439aeaad75e456
MEG3 v1 intermediate form: https://figshare.com/s/a81600774ab9baed8932
MEG3 v1 denatured form: https://figshare.com/s/026d8eb31a8a4b6910e9
MEG3 H11LpA mutant, compact form: https://figshare.com/s/666c9998f22d661d72f3
References
Mattick, J. S. & Rinn, J. L. Discovery and annotation of long noncoding RNAs. Nat. Struct. Mol. Biol. 22, 5–7 (2015).
Ponting, C. P., Oliver, P. L. & Reik, W. Evolution and functions of long noncoding RNAs. Cell 136, 629–641 (2009).
Novikova, I. V., Hennelly, S. P. & Sanbonmatsu, K. Y. Sizing up long non-coding RNAs: do lncRNAs have secondary and tertiary structure? Bioarchitecture 2, 189–199 (2012).
Pyle, A. M. Looking at LncRNAs with the ribozyme toolkit. Mol. Cell 56, 13–17 (2014).
Kaneko, S., Son, J., Bonasio, R., Shen, S. S. & Reinberg, D. Nascent RNA interaction keeps PRC2 activity poised and in check. Genes Dev. 28, 1983–1988 (2014).
Sasaki, Y. T., Ideue, T., Sano, M., Mituyama, T. & Hirose, T. MENepsilon/beta noncoding RNAs are essential for structural integrity of nuclear paraspeckles. Proc. Natl Acad. Sci. USA 106, 2525–2530 (2009).
Schmitt, A. M. et al. An inducible long noncoding RNA amplifies DNA damage signaling. Nat. Genet. 48, 1370–1376 (2016).
Liu, F., Somarowthu, S. & Pyle, A. M. Visualizing the secondary and tertiary architectural domains of lncRNA RepA. Nat. Chem. Biol. 13, 282–289 (2017).
Lu, Z. et al. RNA duplex map in living cells reveals higher-order transcriptome structure. Cell 165, 1267–1279 (2016).
Uroda, T. et al. Conserved pseudoknots in lncRNA MEG3 are essential for stimulation of the p53 pathway. Mol. Cell 75, 982–995.e9 (2019).
Duszczyk, M. M., Wutz, A., Rybin, V. & Sattler, M. The Xist RNA A-repeat comprises a novel AUCG tetraloop fold and a platform for multimerization. RNA 17, 1973–1982 (2011).
Brown, J. A. et al. Structural insights into the stabilization of MALAT1 noncoding RNA by a bipartite triple helix. Nat. Struct. Mol. Biol. 21, 633–640 (2014).
Chillon, I. et al. Native purification and analysis of long RNAs. Methods Enzymol. 558, 3–37 (2015).
Kaneko, S. et al. Interactions between JARID2 and noncoding RNAs regulate PRC2 recruitment to chromatin. Mol. Cell 53, 290–300 (2014).
Zhou, Y. et al. Activation of p53 by MEG3 non-coding RNA. J. Biol. Chem. 282, 24731–24742 (2007).
Feig, A. L. & Uhlenbeck, O. C. in The RNA world (eds R. F. Gesteland, T. R. Cech, & J. F. Atkins) Ch. 12, 287-320 (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, 1999).
Novikova, I. V., Dharap, A., Hennelly, S. P. & Sanbonmatsu, K. Y. 3S: shotgun secondary structure determination of long non-coding RNAs. Methods 63, 170–177 (2013).
Siegfried, N. A., Busan, S., Rice, G. M., Nelson, J. A. & Weeks, K. M. RNA motif discovery by SHAPE and mutational profiling (SHAPE-MaP). Nat. Methods 11, 959–965 (2014).
Volders, P. J. et al. LNCipedia: a database for annotated human lncRNA transcript sequences and structures. Nucleic Acids Res. 41, D246–D251 (2013).
Pennisi, E. Genomics. ENCODE project writes eulogy for junk DNA. Science 337, 1159–1161 (2012).
Novikova, I. V., Hennelly, S. P. & Sanbonmatsu, K. Y. Structural architecture of the human long non-coding RNA, steroid receptor RNA activator. Nucleic Acids Res. 40, 5034–5051 (2012).
Hawkes, E. J. et al. COOLAIR antisense RNAs form evolutionarily conserved elaborate secondary structures. Cell Rep. 16, 3087–3096 (2016).
Lin, Y., Schmidt, B. F., Bruchez, M. P. & McManus, C. J. Structural analyses of NEAT1 lncRNAs suggest long-range RNA interactions that may contribute to paraspeckle architecture. Nucleic Acids Res. 46, 3742–3752 (2018).
Xue, Z. et al. A G-rich motif in the lncRNA Braveheart interacts with a zinc-finger transcription factor to specify the cardiovascular lineage. Mol. Cell 64, 37–50 (2016).
Chillon, I. & Pyle, A. M. Inverted repeat Alu elements in the human lincRNA-p21 adopt a conserved secondary structure that regulates RNA function. Nucleic Acids Res. 44, 9462–9471 (2016).
Somarowthu, S. et al. HOTAIR forms an intricate and modular secondary structure. Mol. Cell 58, 353–361 (2015).
Zhou, Y., Zhang, X. & Klibanski, A. MEG3 noncoding RNA: a tumor suppressor. J. Mol. Endocrinol. 48, R45–R53 (2012).
Smola, M. J. et al. SHAPE reveals transcript-wide interactions, complex structural domains, and protein interactions across the Xist lncRNA in living cells. Proc. Natl Acad. Sci. USA 113, 10322–10327 (2016).
Ilik, I. A. et al. Tandem stem-loops in roX RNAs act together to mediate X chromosome dosage compensation in Drosophila. Mol. Cell 51, 156–173 (2013).
Petrone, J. & DeFrancesco, L. Small molecules get the message. Nat. Biotechnol. 36, 787–790 (2018).
Walder, R. et al. High-precision single-molecule characterization of the folding of an HIV RNA hairpin by atomic force microscopy. Nano Lett. 18, 6318–6325 (2018).
Torelli, E. et al. Isothermal folding of a light-up bio-orthogonal RNA origami nanoribbon. Sci. Rep. 8, 6989 (2018).
Kim, D. N. et al. Zinc-finger protein CNBP alters the 3-D structure of lncRNA Braveheart in solution. Nat. Commun. 11, 148 (2020).
Bustamante, C., Erie, D. A. & Keller, D. Biochemical and structural applications of scanning force microscopy. Curr. Opin. Struct. Biol. 4, 750–760 (1994).
Lyubchenko, Y. L., Shlyakhtenko, L. S. & Ando, T. Imaging of nucleic acids with atomic force microscopy. Methods 54, 274–283 (2011).
Schon, P. Imaging and force probing RNA by atomic force microscopy. Methods 103, 25–33 (2016).
Schon, P. Atomic force microscopy of RNA: state of the art and recent advancements. Semin. Cell Dev. Biol. 73, 209–219 (2018).
Hansma, H. G., Revenko, I., Kim, K. & Laney, D. E. Atomic force microscopy of long and short double-stranded, single-stranded and triple-stranded nucleic acids. Nucleic Acids Res. 24, 713–720 (1996).
Giro, A. et al. Single molecule studies of RNA secondary structure: AFM of TYMV viral RNA. Microsc. Res. Tech. 65, 235–245 (2004).
Garcia-Sacristan, A. et al. A magnesium-induced RNA conformational switch at the internal ribosome entry site of hepatitis C virus genome visualized by atomic force microscopy. Nucleic Acids Res. 43, 565–580 (2015).
Yu, J., Liu, Z., Jiang, W., Wang, G. & Mao, C. De novo design of an RNA tile that self-assembles into a homo-octameric nanoprism. Nat. Commun. 6, 5724 (2015).
Kuhlbrandt, W. Cryo-EM enters a new era. eLife 3, e03678 (2014).
Palovcak, E. et al. A simple and robust procedure for preparing graphene-oxide cryo-EM grids. J. Struct. Biol. 204, 80–84 (2018).
Naydenova, K., Peet, M. J. & Russo, C. J. Multifunctional graphene supports for electron cryomicroscopy. Proc. Natl Acad. Sci. USA 116, 11718–11724 (2019).
Feng, X. et al. A fast and effective microfluidic spraying-plunging method for high-resolution single-particle cryo-EM. Structure 25, 663–670.e3 (2017).
Schmidli, C. et al. Microfluidic protein isolation and sample preparation for high-resolution cryo-EM. Proc. Natl Acad. Sci. USA 116, 15007–15012 (2019).
Miyagi, A. & Scheuring, S. A novel phase-shift-based amplitude detector for a high-speed atomic force microscope. Rev. Sci. Instrum. 89, 083704 (2018).
Trinh, M. H. et al. Tobacco mosaic virus as an AFM tip calibrator. J. Mol. Recognit. 24, 503–510 (2011).
Chen, S. W. et al. Nanoscale structural features determined by AFM for single virus particles. Nanoscale 5, 10877–10886 (2013).
Chen, S. W., Teulon, J. M., Godon, C. & Pellequer, J. L. Atomic force microscope, molecular imaging, and analysis. J. Mol. Recognit. 29, 51–55 (2016).
Godon, C. et al. Conditions to minimize soft single biomolecule deformation when imaging with atomic force microscopy. J. Struct. Biol. 197, 322–329 (2017).
Trinh, M. H. et al. Computational reconstruction of multidomain proteins using atomic force microscopy data. Structure 20, 113–120 (2012).
Niina, T., Fuchigami, S. & Takada, S. Flexible fitting of biomolecular structures to atomic force microscopy images via biased molecular simulations. J. Chem. Theory Comput. 16, 1349–1358 (2020).
Dasgupta, B., Miyashita, O. & Tama, F. Reconstruction of low-resolution molecular structures from simulated atomic force microscopy images. Biochim. Biophys. Acta, Gen. Subj. 1864, 129420 (2020).
Pirakitikulr, N., Kohlway, A., Lindenbach, B. D. & Pyle, A. M. The coding region of the HCV genome contains a network of regulatory RNA structures. Mol. Cell 62, 111–120 (2016).
Mitra, S. Detecting RNA tertiary folding by sedimentation velocity analytical ultracentrifugation. Methods Mol. Biol. 1086, 265–288 (2014).
Garbett, N. C., Mekmaysy, C. S. & Chaires, J. B. Sedimentation velocity ultracentrifugation analysis for hydrodynamic characterization of G-quadruplex structures. Methods Mol. Biol. 608, 97–120 (2010).
Mitra, S. Using analytical ultracentrifugation (AUC) to measure global conformational changes accompanying equilibrium tertiary folding of RNA molecules. Methods Enzymol. 469, 209–236 (2009).
Stetefeld, J., McKenna, S. A. & Patel, T. R. Dynamic light scattering: a practical guide and applications in biomedical sciences. Biophys. Rev. 8, 409–427 (2016).
Patel, T. R. et al. Structural studies of RNA-protein complexes: a hybrid approach involving hydrodynamics, scattering, and computational methods. Methods 118-119, 146–162 (2017).
Chen, Y. & Pollack, L. SAXS studies of RNA: structures, dynamics, and interactions with partners. Wiley Interdiscip. Rev.: RNA 7, 512–526 (2016).
Trewhella, J. Small-angle scattering and 3D structure interpretation. Curr. Opin. Struct. Biol. 40, 1–7 (2016).
Pernot, P. et al. New beamline dedicated to solution scattering from biological macromolecules at the ESRF. J. Phys. Conf. Ser. 247, 012009 (2010).
Press, W. H., Flannery, B. P., Teukolsky, S. A. & Vetterling, W. T. Numerical Recipes in C: The Art of Scientific Computing (Cambridge University Press, 1988).
Petoukhov, M. V. et al. New developments in the ATSAS program package for small-angle scattering data analysis. J. Appl. Crystallogr. 45, 342–350 (2012).
Chen, S. W. & Pellequer, J. L. DeStripe: frequency-based algorithm for removing stripe noises from AFM images. BMC Struct. Biol. 11, 7 (2011).
Necas, D. & Klapetek, P. Gwyddion: an open-source software for SPM data analysis. Cent. Eur. J. Phys. 10, 181–188 (2012).
Delageniere, S. et al. ISPyB: an information management system for synchrotron macromolecular crystallography. Bioinformatics 27, 3186–3192 (2011).
Schuck, P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and Lamm equation modeling. Biophys. J. 78, 1606–1619 (2000).
Zhao, C., Rajashankar, K. R., Marcia, M. & Pyle, A. M. Crystal structure of group II intron domain 1 reveals a template for RNA assembly. Nat. Chem. Biol. 11, 967–972 (2015).
Marcia, M. & Pyle, A. M. Visualizing group II intron catalysis through the stages of splicing. Cell 151, 497–507 (2012).
Li, M. Z. & Elledge, S. J. SLIC: a method for sequence- and ligation-independent cloning. Methods Mol. Biol. 852, 51–59 (2012).
Chaturvedi, S. K., Ma, J., Zhao, H. & Schuck, P. Use of fluorescence-detected sedimentation velocity to study high-affinity protein interactions. Nat. Protoc. 12, 1777–1791 (2017).
Demeler, B. in Modern Analytical Ultracentrifugation: Techniques and Methods (eds Scott, D. J., Harding, S. E. & Rowe, A. J.) 210–229 (Royal Society of Chemistry, 2005).
Su, L. J., Brenowitz, M. & Pyle, A. M. An alternative route for the folding of large RNAs: apparent two-state folding by a group II intron ribozyme. J. Mol. Biol. 334, 639–652 (2003).
Marcia, M. Using molecular replacement phasing to study the structure and function of RNA. Methods Mol. Biol. 1320, 233–257 (2016).
Marcia, M. et al. Solving nucleic acid structures by molecular replacement: examples from group II intron studies. Acta Crystallogr. D. Biol. Crystallogr. 69, 2174–2185 (2013).
Marcia, M. & Pyle, A. M. Principles of ion recognition in RNA: insights from the group II intron structures. RNA 20, 516–527 (2014).
Franke, D. et al. ATSAS 2.8: a comprehensive data analysis suite for small-angle scattering from macromolecular solutions. J. Appl. Crystallogr. 50, 1212–1225 (2017).
Marcia, M., Somarowthu, S. & Pyle, A. M. Now on display: a gallery of group II intron structures at different stages of catalysis. Mob. DNA 4, 14–26 (2013).
Garcia, R. & San Paulo, A. Attractive and repulsive tip-sample interaction regimes in tapping-mode atomic force microscopy. Phys. Rev. B: Condens. Matter 60, 4961–4967 (1999).
Moller, C., Allen, M., Elings, V., Engel, A. & Muller, D. J. Tapping-mode atomic force microscopy produces faithful high-resolution images of protein surfaces. Biophys. J. 77, 1150–1158 (1999).
Teulon, J. M. et al. On the operational aspects of measuring nanoparticle sizes. Nanomaterials (Basel) 9, 18 (2018).
Gilmore, J. L. et al. in Nuclear Bodies and Noncoding RNAs: Methods and Protocols (eds Nakagawa, S. & Hirose, T.) 119–153 (Springer, 2015).
Ando, T. High-speed atomic force microscopy. Curr. Opin. Chem. Biol. 51, 105–112 (2019).
Acknowledgements
We thank A. McCarthy and the beamline scientists at the BM29-BioSAXS beamline for their support during SAXS data collection; A. Leroy and C. Ebel (IBS Grenoble) for support with AUC; S. Acajjaoui and M. Soler Lopez (ESRF Grenoble) for support with DLS; and C. Mas (IBSG Grenoble) and M. Jamin (UGA Grenoble) for support with SEC-MALLS. We also thank all members of the Marcia lab for helpful discussions. Work in the Marcia lab was partly funded by the Agence Nationale de la Recherche (ANR-15-CE11-0003-01), the Agence Nationale de Recherche sur le Sida et les hépatites virales (ANRS, ECTZ18552), ITMO Cancer (18CN047-00), and the Fondation ARC pour la recherche sur le cancer (PJA-20191209284). This work used the platforms of the Grenoble Instruct-ERIC center (ISBG; UMS 3518 CNRS-CEA-UGA-EMBL) within the Grenoble Partnership for Structural Biology (PSB), supported by FRISBI (ANR-10-INBS-05-02) and GRAL, financed within the University Grenoble Alpes graduate school (Ecoles Universitaires de Recherche) CBH-EUR-GS (ANR-17-EURE-0003). IBS acknowledges integration into the Interdisciplinary Research Institute of Grenoble (IRIG, CEA). This work acknowledges the AFM platform at the IBS.
Author information
Authors and Affiliations
Contributions
M.M. conceived the project and obtained funding; M.M. and I.C. supervised the research and wrote the initial draft of the manuscript; T.U., M.M., J.-M.T., and J.-L.P. developed the procedures for AFM image acquisition; T.U., I.C., and M.M. developed the procedures for lncRNA purification, AUC, and MALLS; T.U. and M.M. developed the procedures for DLS and SAXS experiments; M.K. carried out EM experiments; O.P. provided technical assistance and contributed to sample preparation; P.A., J.-L.P., and M.M. developed the procedures for AFM image analysis. All authors reviewed and edited the manuscript and approved the final draft.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Protocols thanks Yuri L. Lyubchenko, Neil H. Thomson and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key reference using this protocol
Uroda, T. et al. Mol. Cell 75, 982–995 (2019): https://doi.org/10.1016/j.molcel.2019.07.025
Extended Data
Extended Data Fig. 1 Negative-staining electron microscopy and atomic force microscopy.
(a) Raw micrograph displaying individual MEG3 particles in 17.5 mM Mg2+ stained with uranyl formate. Aggregates of different size are visible in the image. The scale bar of 100 nm is reported at the top right corner of the panel. (b) Reference-free 2d class averages of single particles selected from the same micrographs as reported in panel A. The scale bar of 30 nm is reported at the top right of the panel. (c) Overview of the mica surface spotted with MEG3 particles in the folded state (scanning resolution = 9.8 nm/pixel). The scale bar of 2 µm is reported at the top right corner of the panel. This panel was adapted from Uroda et al10. (d) Topographical AFM image without particle averaging at a scanning resolution of 0.98 nm/pixel. The scale bar of 400 nm is reported at the top right corner of the panel. This panel is reproduced from Fig. 4, compact form of MEG3.
Extended Data Fig. 2 Cryo-electron microscopy.
Representative micrographs showing MEG3 aggregates as seen on the carbon support of cryo-EM holey grids (in 25 mM Mg2+). The scale bars of 100 nm are reported at the top right corner of each panel.
Extended Data Fig. 3 Denaturing versus non-denaturing purifications of lncRNAs.
(a) SEC-MALLS profiles of MEG3 v1 purified under the non-denaturing conditions described in this protocol. Adapted from Uroda et al10 under a Creative Commons Attribution 4.0 license (https://creativecommons.org/licenses/by/4.0/legalcode). (b) SEC-MALLS of MEG3 v1 purified under denaturing conditions and then refolded using a slow temperature gradient. (c) SEC-MALLS of MEG3 v1 purified under denaturing conditions and then refolded using a fast temperature gradient. (d) DLS profiles of MEG3 v1 purified under non-denaturing conditions or under denaturing conditions and slow refolding. Denaturation and refolding inevitably cause sample heterogeneity.
Extended Data Fig. 4 Hydrodynamic properties of lncRNAs in magnesium-containing buffers.
(a) Representative raw data distribution and residual determined in SEDFIT for MEG3 v1 in 0.1 mM MgCl2. This panel is also represented as an icon in Fig. 1. (b) Sedimentation velocity AUC profiles of MEG3 v1 at the indicated magnesium concentrations. The black arrow indicates signal from a small population of multimeric species or aggregates that form at high magnesium concentrations (> 10 mM). (c) Hill curve representing the Rh distribution at different magnesium concentrations for MEG3 v1. This panel is modified from Uroda et al10. (d) SEC-MALLS profiles of MEG3 v1 eluted in the absence of magnesium and in the presence of 10 mM magnesium. In the latter condition, the aggregates visible by AUC (panel A) compromise the light scattering profile. The panel representing the sample in the absence of magnesium is the same as in Extended Data Fig. 3a. (e) Native agarose gel electrophoresis of MEG3 v1 at the indicated magnesium concentrations (in mM, on the top of each lane). d,e, adapted from Uroda et al10 under a Creative Commons Attribution 4.0 license (https://creativecommons.org/licenses/by/4.0/legalcode).
Extended Data Fig. 5 AFM troubleshooting.
Under suboptimal conditions salt crystals or particle aggregates tend to form on the AFM support. (a) Sample dried too long. The scale bar of 400 nm is reported at the top right corner of the panel. (b) Sample washed insufficiently (50 µL of DEPC-treated water). The scale bar of 400 nm is reported at the top center of the panel. (c) Sample denatured using urea. The scale bar of 400 nm is reported at the top right corner of the panel. (d) Sample deposited on non-derivatized mica (left) vs on mica derivatized with APTES (right). The scale bar of 400 nm is reported at the bottom left corner of each panel.
Extended Data Fig. 6 Quality control of lncRNA purity and homogeneity.
(a) Agarose gel electrophoresis of different transcription reactions of MEG3 v1. M indicate the Quick load 2-log DNA molecular size marker; 1-4 are samples from initial transcription screenings, immediately after transcription (Step 5; 1-4 indicate the transcription buffer used, see Tables 2 and 3); D is a degraded sample; P is a sample transcribed in buffer 4 after SEC (Step 18). The arrows on the right indicate the position of MEG3 and of the linearized transcription vector, which is occasionally visible in samples that have not yet been subjected to DNase treatment, but disappears after purification. (b) Representative Bioanalyzer run of MEG3 v1 transcribed from buffer 4. M is the molecular size marker provided in the Agilent RNA 6000 Nano Kit.
Extended Data Fig. 7 SAXS analysis of lncRNAs.
(a) Scattering curve, (b) Guinier plot, and (c) P(r) plot for MEG3 v1. a–c adapted from Uroda et al10 under a Creative Commons Attribution 4.0 license (https://creativecommons.org/licenses/by/4.0/legalcode). (d) DAMMIF ab initio shape for MEG3 v1. Damaver.pdb output represented as a blue mesh, damfilt.pdb output represented as an orange solid surface (see Step 64 for a description of these files).
Supplementary information
Source data
Source Data Fig. 2
Unprocessed AFM image.
Source Data Fig. 3
Unprocessed AFM image.
Source Data Fig. 4
Unprocessed AFM images.
Source Data Extended Data Fig. 1
Unprocessed AFM and EM images
Source Data Extended Data Fig. 2
Unprocessed EM images.
Source Data Extended Data Fig. 4
Unprocessed agarose gel scans.
Source Data Extended Data Fig. 5
Unprocessed AFM images.
Source Data Extended Data Fig. 6
Unprocessed agarose gel scans.
Rights and permissions
About this article
Cite this article
Uroda, T., Chillón, I., Annibale, P. et al. Visualizing the functional 3D shape and topography of long noncoding RNAs by single-particle atomic force microscopy and in-solution hydrodynamic techniques. Nat Protoc 15, 2107–2139 (2020). https://doi.org/10.1038/s41596-020-0323-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-020-0323-7
This article is cited by
-
RNA structure probing uncovers RNA structure-dependent biological functions
Nature Chemical Biology (2021)
-
iDRiP for the systematic discovery of proteins bound directly to noncoding RNA
Nature Protocols (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.