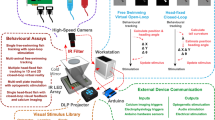
Abstract
Caenorhabditis elegans is a valuable model organism in biomedical research that has led to major discoveries in the fields of neurodegeneration, cancer and aging. Because movement phenotypes are commonly used and represent strong indicators of C. elegans fitness, there is an increasing need to replace manual assessments of worm motility with automated measurements to increase throughput and minimize observer biases. Here, we provide a protocol for the implementation of the improved wide field-of-view nematode tracking platform (WF-NTP), which enables the simultaneous analysis of hundreds of worms with respect to multiple behavioral parameters. The protocol takes only a few hours to complete, excluding the time spent culturing C. elegans, and includes (i) experimental design and preparation of samples, (ii) data recording, (iii) software management with appropriate parameter choices and (iv) post-experimental data analysis. We compare the WF-NTP with other existing worm trackers, including those having high spatial resolution. The main benefits of WF-NTP relate to the high number of worms that can be assessed at the same time on a whole-plate basis and the number of phenotypes that can be screened for simultaneously.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout














Similar content being viewed by others
Data availability
The associated raw data from Figs. 3, 4, 5, 6, 7, 13 and 14 can be accessed via https://doi.org/10.17863/CAM.48480 and demo-data can be accessed via https://doi.org/10.17863/CAM.46983. All other images and movies can be requested via the corresponding authors.
Code availability
The WF-NTP software and plugins can be downloaded from https://github.com/impact27/WF_NTP or https://doi.org/10.5281/zenodo.3630199 (to ensure the version described in this paper: v.3.3.3). We highly recommend downloading the software from GitHub to ensure the latest updates and improvements. The software runs under the license of Attribution-NonCommercial-ShareAlike 4.0 International (CC BY-NC-SA 4.0): https://creativecommons.org/licenses/by-nc-sa/4.0/. The code in this protocol has been peer reviewed.
References
Antoscheckin, I. & Sternberg, P. W. The versatile worm: genetic and genomic resources for Caenorhabditis elegans research. Nat. Rev. Genet. 8, 518–532 (2007).
Hope, I. A. Background on Caenorhabditis elegans. in C. elegans: A Practical Approach (ed Hope I. A.) 1–15 (Oxford University Press, 1999).
Kaletta, T. & Hengartner, M. O. Finding function in novel targets: C. elegans as a model organism. Nat. Rev. Drug Disco. 5, 387–399 (2006).
Leung, M. C. K. et al. Caenorhabditis elegans: an emerging model in biomedical and environmental toxicology. Toxicol. Sci. 106, 5–28 (2008).
Morley, J. F. et al. The threshold for polyglutamine-expansion protein aggregation and cellular toxicity is dynamic and influenced by aging in Caenorhabditis elegans. Proc. Natl Acad. Sci. USA 99, 10417–10422 (2002).
Dillin, A. et al. Rates of behavior and aging specified by mitochondrial function during development. Science 298, 2398–2401 (2002).
Lee, S. S. et al. DAF-16 target genes that control C. elegans life-span and metabolism. Science 300, 644–647 (2003).
Nollen, E. A. A. et al. Genome-wide RNA interference screen identifies previously undescribed regulators of polyglutamine aggregation. Proc. Natl Acad. Sci. USA 101, 6403–6408 (2004).
Kim, Y. & Sun, H. Functional genomic approach to identify novel genes involved in the regulation of oxidative stress resistance and animal lifespan. Aging Cell 6, 489–503 (2007).
Van Ham, T. J. et al. C. elegans model identifies genetic modifiers of α-synuclein inclusion formation during aging. PLoS Genet. 4, e1000027–11 (2008).
Habchi, J. et al. An anticancer drug suppresses the primary nucleation reaction that initiates the production of the toxic Aβ42 aggregates linked with Alzheimer’s disease. Sci. Adv. 2, e1501244 (2016).
Javer, A., Ripoll-Sánchez, L. & Brown, A. E. X. Powerful and interpretable behavioral features for quantitative phenotyping of Caenorhabdities elegans. Philos. Trans. R. Soc. Lond. B Biol. Sci. 373, 20170375 (2018).
Omura, D. T., Clark, D. A., Samuel, A. D. T. & Horvitz, H. R. Dopamine signaling is essential for precise rates of locomotion by C. elegans. PLoS ONE 7, e38649 (2012).
Vidal-Gadea, A. et al. Caenorhabdities elegans selects distinct crawling and swimming gaits via dopamine and serotonin. Proc. Natl Acad. Sci. USA 108, 17504–17509 (2011).
Butler, V. J. et al. A consistent muscle activation strategy underlies crawling and swimming in Caenorhabditis elegans. J. R. Soc. Interface 12, 20140963 (2015).
van Ham, T. J. et al. Identification of MOAG-4/SERF as a regulator of age-related proteotoxicity. Cell 142, 601–612 (2010).
Brignull, H. R., Moore, F. E., Tang, S. J. & Morimoto, R. I. Polyglutamine proteins at the pathogenic threshold display neuron-specific aggregation in a pan-neuronal Caenorhabditis elegans model. J. Neurosci. 26, 7597–7606 (2006).
Ash, P. E. A. et al. Neurotoxic effects of TDP-43 overexpression in C. elegans. Hum. Mol. Genet. 19, 3206–3218 (2010).
Sorrentino, V. et al. Enhancing mitochondrial proteostasis reduces amyloid-β proteotoxicity. Nature 552, 187–193 (2017).
Hahm, J.-H. et al. C. elegans maximum velocity correlates with healthspan and is maintained in worms with an insulin receptor mutation. Nat. Commun. 6, 8919 (2015).
Sleigh, J. N. et al. A novel Caenorhabditis elegans allele, smn-1(cb131), mimicking a mild form of spinal muscular atrophy, provides a convenient drug screening platform highlighting new and pre-approved compounds. Hum. Mol. Genet 20, 245–260 (2010).
Briese, M. et al. Deletion of smn-1, the Caenorhabditis elegans ortholog of the spinal muscular atrophy gene, results in locomotor dysfunction and reduced lifespan. Hum. Mol. Genet 18, 97–104 (2009).
Hewtitt, J. E. et al. Muscle strength deficiency and mitochondrial dysfunction in a muscular dystrophy model of Caenorhabditis elegans and its functional response to drugs. Dis. Model. Mech. 11, dmm036137 (2018).
Wang, J. et al. An ALS-linked mutant SOD1 produces a locomotor defect associated with aggregation and synaptic dysfunction when expressed in neurons of Caenorhabditis elegans. PLoS Genet. 5, e1000350 (2009).
Haroon, S. et al. Multiple molecular mechanisms reduce mtDNA disease in C. elegans. Cell Rep. 22, 3115–3125 (2018).
Park, E. C. & Horvitz, H. R. Mutations with dominant effects on the behavior and morphology of the nematode Caenorhabditis elegans. Genetics 113, 821–852 (1986).
Syntichaki, P. & Tavernarakis, N. Genetic models of mechanotransduction: the nematode Caenorhabdities elegans. Physiol. Rev. 84, 1097–1153 (2004).
Glenn, C. F. et al. Behavioral deficits during early stages of aging in Caenorhabditis elegans result from locomotory deficits possibly linked to muscle frailty. J. Gerontol. A 59, 1251–1260 (2004).
Fang-Yen, C. et al. Biomechanical analysis of gait adaptation in the nematode Caenorhabditis elegans. Proc. Natl Acad. Sci. USA 107, 323–328 (2010).
Karbowski, J. et al. Conservation rules, their breakdown, and optimality in Caenorhabditis sinusoidal locomotion. J. Theor. Biol. 242, 652–669 (2006).
Pierce-Shimomura, J. T. et al. Genetic analysis of crawling and swimming locomotory patterns in C. elegans. Proc. Natl Acad. Sci. USA 105, 20982–20987 (2008).
Korta, J., Clark, D. A., Gabel, C. V., Mahadevan, L. & Samuel, A. D. T. Mechanosensation and mechanical load modulate the locomotory gait of swimming C. elegans. J. Exp. Biol. 210, 2383–2389 (2007).
Houle, D., Govindaraju, D. R. & Omholt, S. Phenomics: the next challenge. Nat. Rev. Genet. 11, 855–866 (2010).
Swierczek, N. A., Giles, A. C., Rankin, C. H. & Kerr, R. A. High-throughput behavioral analysis in C. elegans. Nat. Methods 8, 592–598 (2011).
Perni, M. et al. Massively parallel C. elegans tracking provides multi-dimensional fingerprints for phenotypic discovery. J. Neurosci. Methods 306, 57–67 (2018).
Tsibidis, G. D. & Tavernarakis, N. Nemo: a computational tool for analysing nematode locomotion. BMC Neurosci. 8, 86 (2007).
Stephens, G., Bialek, W. & Ryu, W. S. Dimensionality and dynamics in the behavior of C. elegans. PLoS Comput. Biol. 4, e1000028 (2008).
Brown, A. E. X., Yemini, E. I., Grundy, L. J., Jucikas, T. & Schafer, W. A dictionary of behavioral motifs reveals clusters of genes affecting Caenorhabditis elegans locomotion. Proc. Natl Acad. Sci. USA 110, 791–796 (2013).
Ramot, D., Johnson, B. E., Berry, T. L., Carnell, L. & Goodman, M. B. The parallel worm tracker: a platform for measuring average speed and drug-induced paralysis in nematodes. PLoS ONE 3, e2208 (2008).
Wang, S. J. & Wang, Z.-W. Track-A-Worm, an open-source system for quantitative assessment of C. elegans locomotory and bending behavior. PLoS ONE 8, e69653 (2013).
Stirman, J. N. et al. Real-time multimodel optical control of neurons and muscles in freely behaving Caenorhandities elegans. Nat. Methods 8, 153–158 (2011).
Leifer, A. M., Fang-Yen, C., Hershow, M., Alkema, M. J. & Samuel, A. D. T. Optogenetic manipulation of neural activity in freely moving Caenorhabdities elegans. Nat. Methods 8, 147–152 (2011).
Lee, J. & Park., M. An adaptive background subtraction method based on kernel density estimation. Sens. (Basel) 12, 12279–12300 (2012).
Chiu, S.-Y., Chiu, C.-C., Xu, S., S.-D. A background subtraction algorithm in complex environments based on category entropy analysis. Appl. Sci. 8, 885 (2018).
Allan, D. B., Caswell, T. A. & Keim, N. C. Trackpy v0. 2. ACS Nano 8, 5891–5897 (2014).
Restif, C. et al. CeleST: computer vision software for quantitative analysis of C. elegans swim behavior reveals novel features of locomotion. PLoS Comput. Biol. 17, e1003702 (2014).
Kwon, N., Pyo, J., Lee, S. J. & Je, J. H. 3-D worm tracker for freely moving C. elegans. PLoS ONE 8, e57484 (2013).
Faumont, S. et al. An image-free opto-mechanical system for creating virtual environments and imaging neuronal activity in freely moving Caenorhabditis elegans. PLoS ONE 6, e24666 (2011).
Ghosh, R., Mohammadi, A., Kruglyak, L. & Ryu, W. S. Multiparameter behavioral profiling reveals distinct thermal response regimes in Caenorhabditis elegans. BMC Biol. 10, 85 (2012).
Buckingham, S. D. & Sattelle, D. B. Fast, automated measurement of nematode swimming (thrashing) without morphometry. BMC Neurosci. 10, 84 (2009).
Oh, K. H. & Kim, H. Aldicarb-induced paralysis assay to determine defects in synaptic transmission in Caenorhabdities elgans. Bio. Protoc. 7, e2400 (2017).
Mahoney, T. R., Luo, S. & Nonet, M. L. Analysis of synaptic transmission in Caenorhabditis elgans using an aldicarb-sensitivity assay. Nat. Protoc. 1, 1772–1777 (2006).
Porta-de-la-Riva, M., Fontrodona, L., Villanueva, A. & Cerón, J. Basic Caenorhabditis elegans methods: synchronization and observation. J. Vis. Exp. 10, e4019 (2012).
Koopman, M. et al. A screening-based platform for the assessment of cellular respiration in Caenorhabditis elegans. Nat. Protoc. 11, 1798–1816 (2016).
Mitchell, D. H., Stiles, J. W., Santelli, J. & Sanadi, D. R. Synchronous growth and aging of Caenorhabditis elegans in the presence of fluorodeoxyuridine. J. Gerontol. 34, 28–36 (1979).
Davies, S. K., Leroi, A. M. & Bundy, J. G. Fluorodeoxyuridine affects the identification of metabolic responses to daf-2 status in Caenorhabditis elegans. Mech. Ageing Dev. 133, 46–49 (2012).
Rooney, J. P. et al. Effects of 5ʹ-fluoro-2-deoxyuridine on mitochondrial biology in Caenorhabditis elegans. Exp. Gerontol. 56, 69–76 (2014).
Gruber, J., Ng, L. F., Poovathingsal, S. K. & Halliwell, B. Deceptively simple but simply deceptive – Caenorhabditis elegans lifespan studies: consideration for aging and antioxidant effects. FEBS Lett. 583, 3377–3387 (2009).
Lüersen, K., Faust, U., Gottschling, D.-C. & Döring, F. Gait-specific adaptation of locomotor activity in response to dietary restriction in Caenorhabditis elegans. J. Exp. Biol. 217, 2480–2488 (2014).
van der Goot, A. T. et al. Delaying aging and the aging-associated decline in protein homeostasis by inhibition of tryptophan degradation. Proc. Natl Acad. Sci. USA 109, 14912–14197 (2012).
Zhang, G. et al. A Na+/CL−–coupled GABA transporter, GAT-1, from Caenorhabditis elgans: structural and functional features, specific expression in GABA-ergic neurons, and involvement in muscle function. J. Biol. Chem. 280, 2065–2077 (2005).
Stiernagle, T. Maintenance of C. elegans in WormBook (ed. The C. elegans Research Community) https://doi.org/10.1895/wormbook.1.101.1 (2006).
Fraser, A. et al. Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature 408, 325–330 (2000).
Kamath, R. S. et al. Systematic functional analysis of the C. elegans genome using RNAi. Nature 421, 231–237 (2003).
Faul, F., Erdfelder, E., Lang, A. G. & Buchner, A. G*Power 3: a flexible statistical power analysis program for the social, behavioral, and biomedical sciences. Behav. Res. Methods 39, 175–191 (2007).
Rand, J. B. & Johnson, C. D. Genetic pharmacology: interactions between drugs and gene products in Caenorhabditis elegans. Methods Cell Biol. 48, 187–204 (1995).
Burns, A. R. et al. A predictive model for drug bioaccumulation and bioactivity in Caenorhabditis elegans. Nat. Chem. Biol. 6, 549–557 (2010).
Zheng, S.-Q., Ding, A.-J., Li, G.-P., Wu, G.-S. & Luo, H.-R. Drug absorption efficiency in Caenorhabditis elegans delivered by different methods. PLoS ONE 8, e56877 (2013).
Partridge, F. A., Tearle, A. W., Gravato-Nobre, M. J., Schafer, W. R. & Hodgkin, J. The C. elegans glycosyltransferase BUS-8 has two distinct and essential roles in epidermal morphogenesis. Dev. Biol. 317, 549–559 (2008).
Acknowledgements
We thank the Houtkooper lab for feedback and questions on the WF-NTP software. This project was funded by a European Research Council (ERC) starting grant (281622 PDControl to E.A.A.N.); the Alumni chapter Gooische Groningers, facilitated by the Ubbo Emmius Fonds (to E.A.A.N.); and an Aspasia Fellowship from NWO (015.014.005 to E.A.A.N.).
Author information
Authors and Affiliations
Contributions
M.K. performed most experiments, optimized the final experimental pipeline, adjusted the current WF-NTP software and added new applications and wrote the manuscript together with R.I.S. and E.A.A.N., with contributions from all authors. Q.P. was extensively involved in rewriting, problem solving, editing and optimization of the WF-NTP software. R.I.S. performed experiments on the neurodegenerative strains and helped to verify parameters of the WF-NTP software. M.P., M.V., C.M.D. and T.P.J.K. pioneered the use of the WF-NTP and developed the initial software and platform. M.K., R.I.S. and E.A.A.N. were extensively involved in discussions and interpretations of results.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Protocols thanks Christophe Restif and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key references using this protocol
Limbocker, R. et al. Nat. Commun. 10, 225 (2019): https://doi.org/10.1038/s41467-018-07699-5
Sinnige, T. et al. PLoS ONE 14, e0217746 (2019): https://doi.org/10.1371/journal.pone.0217746
Delivoria, D. C. et al. Sci. Adv. 5, eaax5108 (2019): https://doi.org/10.1126/sciadv.aax5108
Supplementary information
Supplementary Information
Supplementary Tables 1 and 2.
Rights and permissions
About this article
Cite this article
Koopman, M., Peter, Q., Seinstra, R.I. et al. Assessing motor-related phenotypes of Caenorhabditis elegans with the wide field-of-view nematode tracking platform. Nat Protoc 15, 2071–2106 (2020). https://doi.org/10.1038/s41596-020-0321-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-020-0321-9
This article is cited by
-
Skeletonizing Caenorhabditis elegans Based on U-Net Architectures Trained with a Multi-worm Low-Resolution Synthetic Dataset
International Journal of Computer Vision (2023)
-
An economical and highly adaptable optogenetics system for individual and population-level manipulation of Caenorhabditis elegans
BMC Biology (2021)
-
C. elegans: A biosensor for host–microbe interactions
Lab Animal (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.