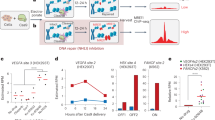
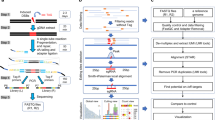
Abstract
DISCOVER-seq (discovery of in situ Cas off-targets and verification by sequencing) is a broadly applicable approach for unbiased CRISPR–Cas off-target identification in cells and tissues. It leverages the recruitment of DNA repair factors to double-strand breaks (DSBs) after genome editing with CRISPR nucleases. Here, we describe a detailed experimental protocol and analysis pipeline with which to perform DISCOVER-seq. The principle of this method is to track the precise recruitment of MRE11 to DSBs by chromatin immunoprecipitation followed by next-generation sequencing. A customized open-source bioinformatics pipeline, BLENDER (blunt end finder), then identifies off-target sequences genome wide. DISCOVER-seq is capable of finding and measuring off-targets in primary cells and in situ. The two main advantages of DISCOVER-seq are (i) low false-positive rates because DNA repair enzyme binding is required for genome edits to occur and (ii) its applicability to a wide variety of systems, including patient-derived cells and animal models. The whole protocol, including the analysis, can be completed within 2 weeks.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Data that support the findings of this study have been deposited in SRA BioProject accession no. PRJNA509652.
Code availability
Our open-source analysis pipeline, called BLENDER (https://github.com/staciawyman/blender), is freely available under the GNU Affero General Public License. The code in this paper has been peer reviewed.
References
Cheng, Y. & Tsai, S. Q. Illuminating the genome-wide activity of genome editors for safe and effective therapeutics. Genome Biol. 19, 226 (2018).
Carroll, D. Collateral damage: benchmarking off-target effects in genome editing. Genome Biol. 20, 114 (2019).
Kleinstiver, B. P. et al. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature 529, 490–495 (2016).
Slaymaker, I. M. et al. Rationally engineered Cas9 nucleases with improved specificity. Science 351, 84–88 (2016).
Vakulskas, C. A. et al. A high-fidelity Cas9 mutant delivered as a ribonucleoprotein complex enables efficient gene editing in human hematopoietic stem and progenitor cells. Nat. Med 24, 1216–1224 (2018).
Chen, J. S. et al. Enhanced proofreading governs CRISPR-Cas9 targeting accuracy. Nature 550, 407–410 (2017).
Wienert, B. et al. Unbiased detection of CRISPR off-targets in vivo using DISCOVER-seq. Science 364, 286–289 (2019).
Stracker, T. H. & Petrini, J. H. J. The MRE11 complex: starting from the ends. Nat. Rev. Mol. Cell Biol. 12, 90–103 (2011).
Williams, R. S., Williams, J. S. & Tainer, J. A. Mre11-Rad50-Nbs1 is a keystone complex connecting DNA repair machinery, double-strand break signaling, and the chromatin template. Biochem. Cell Biol. 85, 509–520 (2007).
Kobayashi, J. Molecular mechanism of the recruitment of NBS1/hMRE11/hRAD50 complex to DNA double-strand breaks: NBS1 binds to gamma-H2AX through FHA/BRCT domain. J. Radiat. Res. 45, 473–478 (2004).
Richardson, C. D. et al. CRISPR-Cas9 genome editing in human cells occurs via the Fanconi anemia pathway. Nat. Genet. 50, 1132–1139 (2018).
Haeussler, M. et al. Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biol. 17, 148 (2016).
Lazzarotto, C. R. et al. Defining CRISPR-Cas9 genome-wide nuclease activities with CIRCLE-seq. Nat. Protoc. 13, 2615–2642 (2018).
Tsai, S. Q. et al. CIRCLE-seq: a highly sensitive in vitro screen for genome-wide CRISPR-Cas9 nuclease off-targets. Nat. Methods 14, 607–614 (2017).
Cameron, P. et al. Mapping the genomic landscape of CRISPR-Cas9 cleavage. Nat. Methods 14, 600–606 (2017).
Kim, D. et al. Digenome-seq: genome-wide profiling of CRISPR-Cas9 off-target effects in human cells. Nat. Methods 12, 237–243 (2015). 1 p following 243.
Tsai, S. Q. et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat. Biotechnol. 33, 187–197 (2015).
Giannoukos, G. et al. UDiTaSTM, a genome editing detection method for indels and genome rearrangements. BMC Genomics 19, 212 (2018).
Frock, R. L. et al. Genome-wide detection of DNA double-stranded breaks induced by engineered nucleases. Nat. Biotechnol. 33, 179–186 (2015).
Crosetto, N. et al. Nucleotide-resolution DNA double-strand break mapping by next-generation sequencing. Nat. Methods 10, 361–365 (2013).
Yan, W. X. et al. BLISS is a versatile and quantitative method for genome-wide profiling of DNA double-strand breaks. Nat. Commun. 8, 15058 (2017).
Akcakaya, P. et al. In vivo CRISPR editing with no detectable genome-wide off-target mutations. Nature 561, 416–419 (2018).
Zetsche, B. et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 163, 759–771 (2015).
Rass, E. et al. Role of Mre11 in chromosomal nonhomologous end joining in mammalian cells. Nat. Struct. Mol. Biol. 16, 819–824 (2009).
Xie, A., Kwok, A. & Scully, R. Role of mammalian Mre11 in classical and alternative nonhomologous end joining. Nat. Struct. Mol. Biol. 16, 814–818 (2009).
Dinkelmann, M. et al. Multiple functions of MRN in end-joining pathways during isotype class switching. Nat. Struct. Mol. Biol. 16, 808–813 (2009).
ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012).
Amemiya, H. M., Kundaje, A. & Boyle, A. P. The ENCODE Blacklist: identification of problematic regions of the genome. Sci. Rep. 9, 9354 (2019).
Wienert, B., Shin, J., Zelin, E., Pestal, K. & Corn, J. E. In vitro-transcribed guide RNAs trigger an innate immune response via the RIG-I pathway. PLoS Biol. 16, e2005840 (2018).
Acknowledgements
J.E.C. and C.D.Y. were supported by the NOMIS Foundation, the Lotte and Adolf Hotz-Sprenger Stiftung, the Swiss National Science Foundation (310030_188858), the Fanconi Anemia Research Foundation, and the Bill and Melinda Gates Foundation. B.W. was supported by an NHMRC Early Career Fellowship. B.R.C and B.W. received support from the Gladstone Institutes and NIH grants R01-EY028249, R01-HL130533 and R01-HL13535801. S.K.W. was supported by the Li Ka Shing Foundation. The authors used the Vincent J. Coates Genomics Sequencing Laboratory at UC Berkeley, which is supported by NIH Instrumentation grant S10 OD018174.
Author information
Authors and Affiliations
Contributions
B.W., S.K.W. and J.E.C. wrote the manuscript with input from all authors. B.W. and S.K.W. developed the original experimental protocol in the J.E.C. lab. C.D.Y. performed ChIP-qPCR time-course experiments. B.W. performed all other experiments. S.K.W. analyzed ChIP-seq data and developed BLENDER software. B.R.C. provided reagents and expertise.
Corresponding authors
Ethics declarations
Competing interests
J.E.C. is a cofounder of Spotlight Therapeutics. J.E.C. has received sponsored research support from AstraZeneca and Pfizer. B.R.C. is a founder of Tenaya Therapeutics.
Additional information
Peer review information Nature Protocols thanks Britta Bouwman, Nicola Crosetto and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related link
Key references using this protocol
Wienert, B. et al. Science 364, 286–289 (2019): https://science.sciencemag.org/content/364/6437/286
Richardson, C. D. et al. Nat. Genet. 50, 1132–1139 (2018): https://www.nature.com/articles/s41588-018-0174-0
Integrated supplementary information
Supplementary Figure 1 Uncropped western blot images.
(a) Whole blot images corresponding to Fig. 2a. Cross-reactivity between human and mouse of five different commercially available MRE11 antibodies as determined by Western Blot. Whole cell extracts (30 μg) from human K562 cells and murine B16-F10 cells were used for the analysis. (b) Whole blot images corresponding to Fig. 2c. Increasing amounts of antibody lead to more MRE11 depletion from cell lysate, as determined by Western Blot. However, increasing pulldown efficiency with larger amounts of antibody is not directly reflected in the levels of eluted MRE11, possibly due to non-linearity of Western blots. FT – flow through, IP - immunoprecipitation.
Supplementary information
Supplementary Information
Supplementary Fig. 1.
Rights and permissions
About this article
Cite this article
Wienert, B., Wyman, S.K., Yeh, C.D. et al. CRISPR off-target detection with DISCOVER-seq. Nat Protoc 15, 1775–1799 (2020). https://doi.org/10.1038/s41596-020-0309-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-020-0309-5
This article is cited by
-
Transient inhibition of 53BP1 increases the frequency of targeted integration in human hematopoietic stem and progenitor cells
Nature Communications (2024)
-
Analysis of Off-target Effects and Risk Assessment Leading from Preclinical to Clinical Trials of Gene-edited Therapeutic Products
Therapeutic Innovation & Regulatory Science (2023)
-
CRISPR in cancer biology and therapy
Nature Reviews Cancer (2022)
-
Frequency and mechanisms of LINE-1 retrotransposon insertions at CRISPR/Cas9 sites
Nature Communications (2022)
-
Cytosine and adenosine base editing in human pluripotent stem cells using transient reporters for editing enrichment
Nature Protocols (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.