Abstract
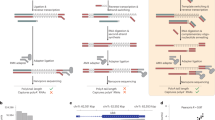
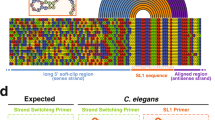
During maturation, eukaryotic precursor RNAs undergo processing events including intron splicing, 3′-end cleavage, and polyadenylation. Here we describe nanopore analysis of co-transcriptional processing (nano-COP), a method for probing the timing and patterns of RNA processing. An extension of native elongating transcript sequencing, which quantifies transcription genome-wide through short-read sequencing of nascent RNA 3′ ends, nano-COP uses long-read nascent RNA sequencing to observe global patterns of RNA processing. First, nascent RNA is stringently purified through a combination of 4-thiouridine metabolic labeling and cellular fractionation. In contrast to cDNA or short-read–based approaches relying on reverse transcription or amplification, the sample is sequenced directly through nanopores to reveal the native context of nascent RNA. nano-COP identifies both active transcription sites and splice isoforms of single RNA molecules during synthesis, providing insight into patterns of intron removal and the physical coupling between transcription and splicing. The nano-COP protocol yields data within 3 d.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The accession numbers for the nanopore sequencing data presented in this paper are Gene Expression Omnibus (GEO): GSE123191 (data from ref. 18) and GSE154079. Supplementary Table 1 indicates the correct accession number for each sample. Source data are provided with this paper.
Code availability
All scripts for data analyses described in this paper are available at https://github.com/churchmanlab/nano-COP. The code for nanopolish-detect-polyI is available at https://github.com/jts/nanopolish.git.
References
Keohavong, P., Gattoni, R., LeMoullec, J. M., Jacob, M. & Stévenin, J. The orderly splicing of the first three leaders of the adenovirus-2 major late transcript. Nucleic Acids Res. 10, 1215–1229 (1982).
Mariman, E. C., van Beek-Reinders, R. J. & van Venrooij, W. J. Alternative splicing pathways exist in the formation of adenoviral late messenger RNAs. J. Mol. Biol. 163, 239–256 (1983).
Beyer, A. L. & Osheim, Y. N. Splice site selection, rate of splicing, and alternative splicing on nascent transcripts. Genes Dev. 2, 754–765 (1988).
Audibert, A., Weil, D. & Dautry, F. In vivo kinetics of mRNA splicing and transport in mammalian cells. Mol. Cell. Biol. 22, 6706–6718 (2002).
Singh, J. & Padgett, R. A. Rates of in situ transcription and splicing in large human genes. Nat. Struct. Mol. Biol. 16, 1128–1133 (2009).
Coulon, A. et al. Kinetic competition during the transcription cycle results in stochastic RNA processing. eLife 3, e03939 (2014).
Martin, R. M., Rino, J., Carvalho, C., Kirchhausen, T. & Carmo-Fonseca, M. Live-cell visualization of pre-mRNA splicing with single-molecule sensitivity. Cell Rep. 4, 1144–1155 (2013).
Rabani, M. et al. High-resolution sequencing and modeling identifies distinct dynamic RNA regulatory strategies. Cell 159, 1698–1710 (2014).
Wachutka, L., Caizzi, L., Gagneur, J. & Cramer, P. Global donor and acceptor splicing site kinetics in human cells. eLife 8, e45056 (2019).
Wan, Y. et al. Dynamic imaging of nascent RNA reveals general principles of transcription dynamics and stochastic splice site selection. SSRN Electron. J. https://doi.org/10.2139/ssrn.3467157 (2019).
Takahara, K. et al. Order of intron removal influences multiple splice outcomes, including a two-exon skip, in a COL5A1 acceptor-site mutation that results in abnormal Pro-α1(V) N-propeptides and ehlers-danlos syndrome type I. Am. J. Hum. Genet. 71, 451–465 (2002).
Fong, N. et al. Pre-mRNA splicing is facilitated by an optimal RNA polymerase II elongation rate. Genes Dev. 28, 2663–2676 (2014).
Dujardin, G. et al. How slow RNA polymerase II elongation favors alternative exon skipping. Mol. Cell 54, 683–690 (2014).
De la Mata, M. et al. A slow RNA polymerase II affects alternative splicing in vivo. Mol. Cell 12, 525–532 (2003).
Kessler, O., Jiang, Y. & Chasin, L. A. Order of intron removal during splicing of endogenous adenine phosphoribosyltransferase and dihydrofolate reductase pre-mRNA. Mol. Cell. Biol. 13, 6211–6222 (1993).
Schwarze, U., Starman, B. J. & Byers, P. H. Redefinition of exon 7 in the COL1A1 gene of type I collagen by an intron 8 splice-donor–site mutation in a form of osteogenesis imperfecta: influence of intron splice order on outcome of splice-site mutation. Am. J. Hum. Genet. 65, 336–344 (1999).
Kim, S. W. et al. Widespread intra-dependencies in the removal of introns from human transcripts. Nucleic Acids Res. 45, 9503–9513 (2017).
Drexler, H. L., Choquet, K. & Churchman, L. S. Splicing kinetics and coordination revealed by direct nascent RNA sequencing through nanopores. Mol. Cell 77, 985–998.e8 (2020).
Churchman, L. S. & Weissman, J. S. Nascent transcript sequencing visualizes transcription at nucleotide resolution. Nature 469, 368–373 (2011).
Mayer, A. et al. Native elongating transcript sequencing reveals human transcriptional activity at nucleotide resolution. Cell 161, 541–554 (2015).
Eid, J. et al. Single polymerase molecules. Science 323, 133–138 (2009).
Weirather, J. L. Et al. Comprehensive comparison of Pacific Biosciences and Oxford Nanopore Technologies and their applications to transcriptome analysis. F1000Res. 6, 100 (2017).
Garalde, D. R. et al. Highly parallel direct RNA sequencing on an array of nanopores. Nat. Methods 15, 201–206 (2018).
Soneson, C. et al. A comprehensive examination of Nanopore native RNA sequencing for characterization of complex transcriptomes. Nat. Commun. 10, 3359 (2019).
Jia, J. et al. Post-transcriptional splicing of nascent RNA contributes to widespread intron retention in plants. Nat. Plants 6, 780–788 (2020).
Danko, C. G. et al. Signaling pathways differentially affect RNA polymerase II initiation, pausing, and elongation rate in cells. Mol. Cell 50, 212–222 (2013).
Veloso, A. et al. Rate of elongation by RNA polymerase II is associated with specific gene features and epigenetic modifications. Genome Res. 24, 896–905 (2014).
Dölken, L. et al. High-resolution gene expression profiling for simultaneous kinetic parameter analysis of RNA synthesis and decay. RNA 14, 1959–1972 (2008).
Windhager, L. et al. Ultrashort and progressive 4sU-tagging reveals key characteristics of RNA processing at nucleotide resolution. Genome Res. 22, 2031–2042 (2012).
Schwalb, B. et al. TT-seq maps the human transient transcriptome. Science 352, 1225–1228 (2016).
Rabani, M. et al. Metabolic labeling of RNA uncovers principles of RNA production and degradation dynamics in mammalian cells. Nat. Biotechnol. 29, 436–442 (2011).
Gregersen, L. H., Mitter, R. & Svejstrup, J. Q. Using Ttchem-seq for profiling nascent transcription and measuring transcript elongation. Nat. Protoc. 15, 604–627 (2020).
Pai, A. A. et al. The kinetics of pre-mRNA splicing in the Drosophila genome and the influence of gene architecture. eLife 6, 1–26 (2017).
Maier, K. C., Gressel, S., Cramer, P. & Schwalb, B. Native molecule sequencing by nano-ID reveals synthesis and stability of RNA isoforms. Genome Res. 30, 1332–1344 (2020).
Carrillo Oesterreich, F. et al. Splicing of nascent RNA coincides with intron exit from RNA polymerase II. Cell 165, 372–381 (2016).
Brody, Y. et al. The In vivo kinetics of RNA polymerase II elongation during co-transcriptional splicing. PloS Biol. 9, e1000573 (2011).
Herzel, L., Straube, K. & Neugebauer, K. M. Long-read sequencing of nascent RNA reveals coupling among RNA processing events. Genome Res. 28, 1008–1019 (2018).
Mayer, A. & Churchman, L. S. Genome-wide profiling of RNA polymerase transcription at nucleotide resolution in human cells with native elongating transcript sequencing. Nat. Protoc. 11, 813–833 (2016).
Lindell, T. J., Weinberg, F., Morris, P. W., Roeder, R. G. & Rutter, W. J. Specific inhibition of nuclear RNA polymerase II by alpha-amanitin. Science 170, 447–449 (1970).
Li, H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34, 3094–3100 (2018).
Han, F. & Lillard, S. J. In-situ sampling and separation of RNA from individual mammalian cells. Anal. Chem. 72, 4073–4079 (2000).
Jackson, D. A., Iborra, F. J., Manders, E. M. & Cook, P. R. Numbers and organization of RNA polymerases, nascent transcripts, and transcription units in HeLa nuclei. Mol. Biol. Cell 9, 1523–1536 (1998).
Rädle, B. et al. Metabolic labeling of newly transcribed RNA for high resolution gene expression profiling of RNA synthesis, processing and decay in cell culture. J. Vis. Exp. https://doi.org/10.3791/50195 (2013).
Tani, H. et al. Genome-wide determination of RNA stability reveals hundreds of short-lived noncoding transcripts in mammals. Genome Res. 22, 947–956 (2012).
Schofield, J. A., Duffy, E. E., Kiefer, L., Sullivan, M. C. & Simon, M. D. TimeLapse-seq: adding a temporal dimension to RNA sequencing through nucleoside recoding. Nat. Methods 15, 221–225 (2018).
Kovaka, S., Fan, Y., Ni, B., Timp, W. & Schatz, M. C. Targeted nanopore sequencing by real-time mapping of raw electrical signal with UNCALLED. Nat. Biotechnol. https://doi.org/10.1038/s41587-020-0731-9 (2020).
Payne, A. et al. Nanopore adaptive sequencing for mixed samples, whole exome capture and targeted panels. Preprint at bioRxiv https://doi.org/10.1101/2020.02.03.926956 (2020).
Li, H. et al. The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Dale, R. K., Pedersen, B. S. & Quinlan, A. R. Pybedtools: a flexible Python library for manipulating genomic datasets and annotations. Bioinformatics 27, 3423–3424 (2011).
Workman, R. E. et al. Nanopore native RNA sequencing of a human poly(A) transcriptome. Nat. Methods 16, 1297–1305 (2019).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
The ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012).
Acknowledgements
We thank members of the Churchman lab, F. Winston, W. Timp, R. Workman, N. Sadowski, M. Marin, B. Smalec, M. Richardson, R. Ietswaart, and A. Markham for helpful discussions, advice, and assistance and C. Kaplan, J. Bridgers, B. Smalec, J. Falk and C. Patil for critical reading of the manuscript. This work was supported by the NIH (R21-HG009264, R01-HG010538, and R01-GM117333 to L.S.C.; F31-GM122133 to H.L.D.), an NSF Graduate Research Fellowship to H.E.M., the Fonds de Recherche du Québec–Santé and the Canadian Institutes of Health Research (Post-doctoral fellowship awards to K.C.). J.T.S. is supported by the Ontario Institute for Cancer Research through funds provided by the Government of Ontario and the Government of Canada through Genome Canada and Ontario Genomics (OGI-136).
Author information
Authors and Affiliations
Contributions
H.L.D., K.C. and L.S.C. conceived and designed the study. H.L.D. established the nano-COP protocol and K.C. developed the poly(I) tailing approach. H.L.D., K.C. and H.E.M. performed experiments. H.L.D. and K.C. generated scripts and performed data analysis. P.S.T. and J.T.S. developed nanopolish-detect-polyI. H.L.D., K.C., H.E.M. and L.S.C. wrote the manuscript. P.S.T. and J.T.S. reviewed and edited the manuscript.
Corresponding author
Ethics declarations
Competing interests
J.T.S. receives research funding from Oxford Nanopore Technologies. J.T.S. and H.L.D. have received travel support to attend and speak at meetings organized by Oxford Nanopore Technologies. All other authors declare no competing interests.
Additional information
Peer review information Nature Protocols thanks Shobbir Hussain, Jixian Zhai and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key reference using this protocol:
Drexler, H. L., Choquet, K. & Churchman, L. S. Mol. Cell 77, 985–998.e8 (2020): https://doi.org/10.1016/j.molcel.2019.11.017
This protocol is an extension to: Nat. Protoc. 11, 813–833 (2016): https://doi.org/10.1038/nprot.2016.047
Supplementary information
Supplementary Information
Supplementary Methods, Supplementary Fig. 1, Supplementary Note and Supplementary Table 1
Source data
Source Data Fig. 3
Unprocessed gel.
Rights and permissions
About this article
Cite this article
Drexler, H.L., Choquet, K., Merens, H.E. et al. Revealing nascent RNA processing dynamics with nano-COP. Nat Protoc 16, 1343–1375 (2021). https://doi.org/10.1038/s41596-020-00469-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-020-00469-y
This article is cited by
-
Co-transcriptional gene regulation in eukaryotes and prokaryotes
Nature Reviews Molecular Cell Biology (2024)
-
Pre-mRNA splicing order is predetermined and maintains splicing fidelity across multi-intronic transcripts
Nature Structural & Molecular Biology (2023)
-
High-sensitive nascent transcript sequencing reveals BRD4-specific control of widespread enhancer and target gene transcription
Nature Communications (2023)
-
Approaching complete genomes, transcriptomes and epi-omes with accurate long-read sequencing
Nature Methods (2023)
-
Time-resolved single-cell RNA-seq using metabolic RNA labelling
Nature Reviews Methods Primers (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.