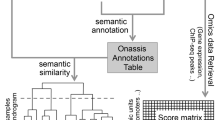
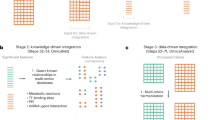
Abstract
Cognitive computing is revolutionizing the way big data are processed and integrated, with artificial intelligence (AI) natural language processing (NLP) platforms helping researchers to efficiently search and digest the vast scientific literature. Most available platforms have been developed for biomedical researchers, but new NLP tools are emerging for biologists in other fields and an important example is metabolomics. NLP provides literature-based contextualization of metabolic features that decreases the time and expert-level subject knowledge required during the prioritization, identification and interpretation steps in the metabolomics data analysis pipeline. Here, we describe and demonstrate four workflows that combine metabolomics data with NLP-based literature searches of scientific databases to aid in the analysis of metabolomics data and their biological interpretation. The four procedures can be used in isolation or consecutively, depending on the research questions. The first, used for initial metabolite annotation and prioritization, creates a list of metabolites that would be interesting for follow-up. The second workflow finds literature evidence of the activity of metabolites and metabolic pathways in governing the biological condition on a systems biology level. The third is used to identify candidate biomarkers, and the fourth looks for metabolic conditions or drug-repurposing targets that the two diseases have in common. The protocol can take 1–4 h or more to complete, depending on the processing time of the various software used.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout















Similar content being viewed by others
Data availability
The datasets analyzed during the current study that were not generated by the authors but mined from public sources are available in the MetaboLights repository (MTBLS298), the Human Metabolome Database (https://hmdb.ca/unearth/q?utf8=%E2%9C%93&query=NAFLD&searcher=diseases&button=), or the main text and Supplementary Information, or upon request of the author of the following publications: ‘Metabolomics identifies perturbations in human disorders of propionate metabolism’ (https://doi.org/10.1373/clinchem.2007.089011), ‘Metabolism links bacterial biofilms and colon carcinogenesis’ (https://doi.org/10.1016/j.cmet.2015.04.011) and ‘Systems biology guided by XCMS Online metabolomics’ (https://doi.org/10.1038/nmeth.4260). Additional data generated by the authors or analyzed during this study are included in this published article and its Supplementary Information files.
References
Kurczy, M. E. et al. Determining conserved metabolic biomarkers from a million database queries. Bioinformatics 31, 3721–3724 (2015).
Monteiro, M. S., Carvalho, M., Bastos, M. L. & Guedes de Pinho, P. Metabolomics analysis for biomarker discovery: advances and challenges. Curr. Med. Chem. 20, 257–271 (2013).
Zhang, A., Sun, H., Yan, G., Wang, P. & Wang, X. Metabolomics for biomarker discovery: moving to the clinic. Biomed. Res. Int. 2015, 354671–354671 (2015).
Zhang, F. et al. Metabolomics for biomarker discovery in the diagnosis, prognosis, survival and recurrence of colorectal cancer: a systematic review. Oncotarget 8, 35460–35472 (2017).
Taylor, J., King, R. D., Altmann, T. & Fiehn, O. Application of metabolomics to plant genotype discrimination using statistics and machine learning. Bioinformatics 18, S241–S248 (2002).
Guijas, C., Montenegro-Burke, J. R., Warth, B., Spilker, M. E. & Siuzdak, G. Metabolomics activity screening for identifying metabolites that modulate phenotype. Nat. Biotechnol. 36, 316–320 (2018).
Paris, L. P. et al. Global metabolomics reveals metabolic dysregulation in ischemic retinopathy. Metabolomics 12, 15 (2016).
Goodacre, R. et al. Proposed minimum reporting standards for data analysis in metabolomics. Metabolomics 3, 231–241 (2007).
Guijas, C. et al. METLIN: a technology platform for identifying knowns and unknowns. Anal. Chem. 90, 3156–3164 (2018).
Domingo-Almenara, X., Montenegro-Burke, J. R., Benton, H. P. & Siuzdak, G. Annotation: a computational solution for streamlining metabolomics analysis. Anal. Chem. 90, 480–489 (2018).
Smolinska, A. et al. Current breathomics—a review on data pre-processing techniques and machine learning in metabolomics breath analysis. J. Breath Res. 8, 027105 (2014).
Kell, D. B. Metabolomics and systems biology: making sense of the soup. Curr. Opin. Microbiol. 7, 296–307 (2004).
Spasić, I. et al. MeMo: a hybrid SQL/XML approach to metabolomic data management for functional genomics. BMC Bioinf. 7, 281 (2006).
Spasić, I. et al. Facilitating the development of controlled vocabularies for metabolomics technologies with text mining. BMC Bioinf. 9, S5 (2008).
Tenopir, C., King, D. W., Christian, L. & Volentine, R. Scholarly article seeking, reading, and use: a continuing evolution from print to electronic in the sciences and social sciences. Learned Publ. 28, 93–105 (2015).
Bornmann, L. & Mutz, R. Growth rates of modern science: a bibliometric analysis based on the number of publications and cited references. J. Assoc. Inf. Sci. Technol. 66, 2215–2222 (2015).
de Solla Price, D. J. Networks of scientific papers. Science 149, 510–515 (1965).
Yandell, M. D. & Majoros, W. H. Genomics and natural language processing. Nat. Rev. Genet. 3, 601–610 (2002).
Hirschberg, J. & Manning, C. D. Advances in natural language processing. Science 349, 261–266 (2015).
Chen, Y., Elenee Argentinis, J. D. & Weber, G. IBM Watson: how cognitive computing can be applied to big data challenges in life sciences research. Clin. Ther. 38, 688–701 (2016).
Choi, B.-K. et al. Literature-based automated discovery of tumor suppressor p53 phosphorylation and inhibition by NEK2. Proc. Natl. Acad. Sci. USA 115, 10666–10671 (2018).
Bakkar, N. et al. Artificial intelligence in neurodegenerative disease research: use of IBM Watson to identify additional RNA-binding proteins altered in amyotrophic lateral sclerosis. Acta Neuropathol. 135, 227–247 (2018).
Ivanisevic, J. et al. Toward ‘omic scale metabolite profiling: a dual separation–mass spectrometry approach for coverage of lipid and central carbon metabolism. Anal. Chem. 85, 6876–6884 (2013).
Forsberg, E. M. et al. Data processing, multi-omic pathway mapping, and metabolite activity analysis using XCMS Online. Nat. Protoc. 13, 633–651 (2018).
Zhu, Z.-J. et al. Liquid chromatography quadrupole time-of-flight mass spectrometry characterization of metabolites guided by the METLIN database. Nat. Protoc. 8, 451–460 (2013).
Patti, G. J., Tautenhahn, R. & Siuzdak, G. Meta-analysis of untargeted metabolomic data from multiple profiling experiments. Nat. Protoc. 7, 508–516 (2012).
Domingo-Almenara, X. et al. XCMS-MRM and METLIN-MRM: a cloud library and public resource for targeted analysis of small molecules. Nat. Methods 15, 681–684 (2018).
Smith, C. A., Want, E. J., O’Maille, G., Abagyan, R. & Siuzdak, G. XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal. Chem. 78, 779–787 (2006).
Tautenhahn, R., Patti, G. J., Rinehart, D. & Siuzdak, G. XCMS Online: a web-based platform to process untargeted metabolomic data. Anal. Chem. 84, 5035–5039 (2012).
Bhinderwala, F. & Powers, R. NMR metabolomics protocols for drug discovery. Methods Mol. Biol. 2037, 265–311 (2019).
Bliziotis, N. G. et al. A comparison of high-throughput plasma NMR protocols for comparative untargeted metabolomics. Metabolomics 16, 64 (2020).
Das, S., Edison, A. S. & Merz, K. M. Metabolite structure assignment using in silico NMR techniques. Anal. Chem. 92, 10412–10419 (2020).
Divaris, K. et al. The supragingival biofilm in early childhood caries: clinical and laboratory protocols and bioinformatics pipelines supporting metagenomics, metatranscriptomics, and metabolomics studies of the oral microbiome. Methods Mol. Biol. 1922, 525–548 (2019).
Erban, A. et al. Multiplexed profiling and data processing methods to identify temperature-regulated primary metabolites using gas chromatography coupled to mass spectrometry. Methods Mol. Biol. 2156, 203–239 (2020).
Palmas, F., Mussap, M. & Fattuoni, C. Urine metabolome analysis by gas chromatography-mass spectrometry (GC-MS): standardization and optimization of protocols for urea removal and short-term sample storage. Clin. Chim. Acta 485, 236–242 (2018).
Papadimitropoulos, M. P., Vasilopoulou, C. G., Maga-Nteve, C. & Klapa, M. I. Untargeted GC-MS metabolomics. Methods Mol. Biol. 1738, 133–147 (2018).
Zarate, E. et al. Fully automated trimethylsilyl (tms) derivatisation protocol for metabolite profiling by GC-MS. Metabolites 7, 1 (2016).
Huan, T. et al. Systems biology guided by XCMS Online metabolomics. Nat. Methods 14, 461–462 (2017).
Gowda, H. et al. Interactive XCMS Online: simplifying advanced metabolomic data processing and subsequent statistical analyses. Anal. Chem. 86, 6931–6939 (2014).
Chong, J. et al. MetaboAnalyst 4.0: towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 46, W486–W494 (2018).
Chong, J., Yamamoto, M. & Xia, J. MetaboAnalystR 2.0: from raw spectra to biological insights. Metabolites 9, 57 (2019).
Tsugawa, H. et al. MS-DIAL: data-independent MS/MS deconvolution for comprehensive metabolome analysis. Nat. Methods 12, 523–526 (2015).
Pluskal, T., Castillo, S., Villar-Briones, A. & Oresic, M. MZmine 2: modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinf. 11, 395 (2010).
O’Shea, K. & Misra, B. B. Software tools, databases and resources in metabolomics: updates from 2018 to 2019. Metabolomics 16, 36 (2020).
Domingo-Almenara, X. et al. Autonomous METLIN-guided in-source fragment annotation for untargeted metabolomics. Anal. Chem. 91, 3246–3253 (2019).
Chong, J., Wishart, D. S. & Xia, J. Using MetaboAnalyst 4.0 for comprehensive and integrative metabolomics data analysis. Curr. Protoc. Bioinformatics 68, e86 (2019).
Chong, J. & Xia, J. Using MetaboAnalyst 4.0 for metabolomics data analysis, interpretation, and integration with other omics data. Methods Mol. Biol. 2104, 337–360 (2020).
Blaženović, I., Kind, T., Ji, J. & Fiehn, O. Software tools and approaches for compound identification of LC-MS/MS data in metabolomics. Metabolites 8, 31 (2018).
Gabrielson, S. W. SciFinder. JMLA 106, 588–590 (2018).
Yu, K.-H. et al. A cloud-based metabolite and chemical prioritization system for the biology/disease-driven Human Proteome Project. J. Proteome Res. 17, 4345–4357 (2018).
Johnston, T. H. et al. Repurposing drugs to treat l-DOPA-induced dyskinesia in Parkinson’s disease. Neuropharmacology 147, 11–27 (2018).
Warth, B. et al. Exposome-scale investigations guided by global metabolomics, pathway analysis, and cognitive computing. Anal. Chem. 89, 11505–11513 (2017).
Guijas, C. et al. Metabolic adaptation to calorie restriction. Sci. Signaling 13, eabb2490 (2020).
Rinschen, M. M. et al. Metabolic rewiring of the hypertensive kidney. Sci. Signaling 12, eaax9760 (2019).
Rey, F. E. et al. Metabolic niche of a prominent sulfate-reducing human gut bacterium. Proc. Natl. Acad. Sci. USA 110, 13582–13587 (2013).
Junping, Z. et al. N‐Acetyl‐cysteine alleviates gut dysbiosis and glucose metabolic disorder in high‐fat diet‐induced mice. J. Diabetes 11, 32–45 (2019).
Hale, V. L. et al. Synthesis of multi-omic data and community metabolic models reveals insights into the role of hydrogen sulfide in colon cancer. Methods 149, 59–68 (2018).
Hyötyläinen, T. et al. Genome-scale study reveals reduced metabolic adaptability in patients with non-alcoholic fatty liver disease. Nat. Commun. 7, 8994 (2016).
Raman, M. et al. Fecal microbiome and volatile organic compound metabolome in obese humans with nonalcoholic fatty liver disease. Clin. Gastroenterol. Hepatol. 11, 868–875.e863 (2013).
Scheller, R. et al. Toward mechanistic models for genotype–phenotype correlations in phenylketonuria using protein stability calculations. Hum. Mutat. 40, 444–457 (2019).
Chen, T. et al. Mutational and phenotypic spectrum of phenylalanine hydroxylase deficiency in Zhejiang Province, China. Sci. Rep. 8, 17137 (2018).
Duan, H. et al. Non-invasive prenatal testing of pregnancies at risk for phenylketonuria. Arch. Dis. Child. Fetal Neonatal Ed. 104, F24–F29 (2019).
Zori, R. et al. Induction, titration, and maintenance dosing regimen in a phase 2 study of pegvaliase for control of blood phenylalanine in adults with phenylketonuria. Mol. Genet. Metab. 125, 217–227 (2018).
Brantley, K. D., Douglas, T. D. & Singh, R. H. One-year follow-up of B vitamin and iron status in patients with phenylketonuria provided tetrahydrobiopterin (BH4). Orphanet J. Rare Dis. 13, 192 (2018).
Johnson, C. H., Ivanisevic, J. & Siuzdak, G. Metabolomics: beyond biomarkers and towards mechanisms. Nat. Rev. Mol. Cell Biol. 17, 451–459 (2016).
Wikoff, W. R., Gangoiti, J. A., Barshop, B. A. & Siuzdak, G. Metabolomics identifies perturbations in human disorders of propionate metabolism. Clin. Chem. 53, 2169–2176 (2007).
Kenny, L. C. et al. Novel biomarkers for pre-eclampsia detected using metabolomics and machine learning. Metabolomics 1, 227 (2005).
Go, A. S., Chertow, G. M., Fan, D., McCulloch, C. E. & Hsu, C.-Y. Chronic kidney disease and the risks of death, cardiovascular events, and hospitalization. N. Engl. J. Med. 351, 1296–1305 (2004).
Tuñón, J. et al. Design and rationale of a multicentre, randomised, double-blind, placebo-controlled clinical trial to evaluate the effect of vitamin D on ventricular remodelling in patients with anterior myocardial infarction: the VITamin D in Acute Myocardial Infarction (VITDAMI) trial. BMJ Open 6, e011287 (2016).
Fricke, S. Semantic Scholar. JMLA 106, 145–147 (2018).
Toonen, L. J. A. et al. Transcriptional profiling and biomarker identification reveal tissue specific effects of expanded ataxin-3 in a spinocerebellar ataxia type 3 mouse model. Mol. Neurodegener. 13, 31 (2018).
Roden, D. & Denny, J. Integrating electronic health record genotype and phenotype datasets to transform patient care. Clin. Pharmacol. Ther. 99, 298–305 (2016).
Krittanawong, C., Zhang, H., Wang, Z., Aydar, M. & Kitai, T. Artificial intelligence in precision cardiovascular medicine. J. Am. Coll. Cardiol. 69, 2657–2664 (2017).
Palmblad, M. Visual and semantic enrichment of analytical chemistry literature searches by combining text mining and computational chemistry. Anal. Chem. 91, 4312–4316 (2019).
Venkatesan, A. et al. SciLite: a platform for displaying text-mined annotations as a means to link research articles with biological data [version 2; referees: 2 approved, 1 approved with reservations]. Wellcome Open Res. https://doi.org/10.12688/wellcomeopenres.10210.2 (2017).
Soto, A. J., Przybyła, P. & Ananiadou, S. Thalia: semantic search engine for biomedical abstracts. Bioinformatics 35, 1799–1801 (2018).
Miwa, M., Thompson, P. & Ananiadou, S. Boosting automatic event extraction from the literature using domain adaptation and coreference resolution. Bioinformatics 28, 1759–1765 (2012).
Nobata, C. et al. Mining metabolites: extracting the yeast metabolome from the literature. Metabolomics 7, 94–101 (2011).
Wei, C. H., Kao, H. Y. & Lu, Z. PubTator: a web-based text mining tool for assisting biocuration. Nucleic Acids Res. 41, W518–W522 (2013).
Wei, C. H., Allot, A., Leaman, R. & Lu, Z. PubTator central: automated concept annotation for biomedical full text articles. Nucleic Acids Res. 47, W587–W593 (2019).
Mohimani, H. et al. Dereplication of microbial metabolites through database search of mass spectra. Nat. Commun. 9, 4035 (2018).
Lai, Z. et al. Identifying metabolites by integrating metabolome databases with mass spectrometry cheminformatics. Nat. Methods 15, 53–56 (2018).
Tsugawa, H. et al. A cheminformatics approach to characterize metabolomes in stable-isotope-labeled organisms. Nat. Methods 16, 295–298 (2019).
Pence, H. E. & Williams, A. ChemSpider: an online chemical information resource. J. Chem. Educ. 87, 1123–1124 (2010).
Heinonen, M., Shen, H., Zamboni, N. & Rousu, J. Metabolite identification and molecular fingerprint prediction through machine learning. Bioinformatics 28, 2333–2341 (2012).
Kim, S. et al. Literature information in PubChem: associations between PubChem records and scientific articles. J. Cheminform 8, 32 (2016).
Wishart, D. S. et al. HMDB 4.0: the human metabolome database for 2018. Nucleic Acids Res. 46, D608–D617 (2018).
Ramirez-Gaona, M. et al. YMDB 2.0: a significantly expanded version of the yeast metabolome database. Nucleic Acids Res. 45, D440–D445 (2017).
Kanehisa, M., Sato, Y., Kawashima, M., Furumichi, M. & Tanabe, M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 44, D457–D462 (2016).
Kanehisa, M. KEGG bioinformatics resource for plant genomics and metabolomics. Methods Mol. Biol. 1374, 55–70 (2016).
Kanehisa, M., Furumichi, M., Tanabe, M., Sato, Y. & Morishima, K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 45, D353–D361 (2017).
Szklarczyk, D. et al. STITCH 5: augmenting protein-chemical interaction networks with tissue and affinity data. Nucleic Acids Res. 44, D380–D384 (2016).
The UniProt Consortium. UniProt: a hub for protein information. Nucleic Acids Res. 43, D204–D212 (2015).
Adams, K. J. et al. Skyline for Small Molecules: a unifying software package for quantitative metabolomics. J. Proteome Res. 19, 1447–1458 (2020).
Zukunft, S. et al. High-throughput extraction and quantification method for targeted metabolomics in murine tissues. Metabolomics 14, 18 (2018).
Yang, B., Tsui, T., Caprioli, R. M. & Norris, J. L. Sample preparation and analysis of single cells using high performance MALDI FTICR mass spectrometry. Methods Mol. Biol. 2064, 125–134 (2020).
Maia, M. et al. Metabolite extraction for high-throughput FTICR-MS-based metabolomics of grapevine leaves. EuPA Open Proteom. 12, 4–9 (2016).
Southam, A. D., Weber, R. J., Engel, J., Jones, M. R. & Viant, M. R. A complete workflow for high-resolution spectral-stitching nanoelectrospray direct-infusion mass-spectrometry-based metabolomics and lipidomics. Nat. Protoc. 12, 310–328 (2016).
Snytnikova, O. A., Khlichkina, A. A., Sagdeev, R. Z. & Tsentalovich, Y. P. Evaluation of sample preparation protocols for quantitative NMR-based metabolomics. Metabolomics 15, 84 (2019).
Spicer, R., Salek, R. M., Moreno, P., Cañueto, D. & Steinbeck, C. Navigating freely-available software tools for metabolomics analysis. Metabolomics 13, 106 (2017).
Sansone, S.-A. et al. Metabolomics standards initiative: ontology working group work in progress. Metabolomics 3, 249–256 (2007).
Sumner, L. W. et al. Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics 3, 211–221 (2007).
Eicher, T. et al. Metabolomics and multi-omics integration: a survey of computational methods and resources. Metabolites 10, 202 (2020).
Misra, B. B. Open-source software tools, databases, and resources for single-cell and single-cell-type metabolomics. Methods Mol. Biol. 2064, 191–217 (2020).
Caspi, R. et al. The MetaCyc database of metabolic pathways and enzymes. Nucleic Acids Res. 46, D633–D639 (2017).
Wishart, D. S. et al. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 46, D1074–D1082 (2018).
Johnson, C. H. et al. Metabolism links bacterial biofilms and colon carcinogenesis. Cell Metab. 21, 891–897 (2015).
Beyer, B. A. et al. Metabolomics-based discovery of a metabolite that enhances oligodendrocyte maturation. Nat. Chem. Biol. 14, 22 (2017).
Wolswijk, G. Oligodendrocyte precursor cells in the demyelinated multiple sclerosis spinal cord. Brain 125, 338–349 (2002).
Boelen, A. et al. Type 3 deiodinase expression in inflammatory spinal cord lesions in rat experimental autoimmune encephalomyelitis. Thyroid 19, 1401–1406 (2009).
Gallai, V. et al. Neuropeptide Y plasma levels and serum dopamine-beta-hydroxylase activity in MS patients with and without abnormal cardiovascular reflexes. Acta Neurol. Belg. 94, 44–52 (1994).
Mann, M. B. et al. Association between the phenylethanolamine N-methyltransferase gene and multiple sclerosis. J. Neuroimmunol. 124, 101–105 (2002).
Cosentino, M. et al. Catecholamine production and tyrosine hydroxylase expression in peripheral blood mononuclear cells from multiple sclerosis patients: effect of cell stimulation and possible relevance for activation-induced apoptosis. J. Neuroimmunol. 133, 233–240 (2002).
Niland, B. et al. Cleavage of transaldolase by granzyme B causes the loss of enzymatic activity with retention of antigenicity for multiple sclerosis patients. J. Immunol. 184, 4025–4032 (2010).
Samland, A. K. & Sprenger, G. A. Transaldolase: from biochemistry to human disease. Int. J. Biochem. Cell Biol. 41, 1482–1494 (2009).
Esposito, M. et al. Human transaldolase and cross-reactive viral epitopes identified by autoantibodies of multiple sclerosis patients. J. Immunol. 163, 4027–4032 (1999).
Banki, K. et al. Oligodendrocyte-specific expression and autoantigenicity of transaldolase in multiple sclerosis. J. Exp. Med. 180, 1649–1663 (1994).
Dousset, J.-C., Trouilh, M. & Foglietti, M.-J. Plasma malonaldehyde levels during myocardial infarction. Clin. Chim. Acta 129, 319–322 (1983).
Loughrey, C. M. et al. Oxidative stress in haemodialysis. QJM 87, 679–683 (1994).
Lim, C. S. & Vaziri, N. D. The effects of iron dextran on the oxidative stress in cardiovascular tissues of rats with chronic renal failure. Kidney Int. 65, 1802–1809 (2004).
Virella, G. & Lopes-Virella, M. F. The pathogenic role of the adaptive immune response to modified LDL in diabetes. Front. Endocrinol. (Lausanne) 3, 76 (2012).
Vallejo, J., Duner, P., Fredrikson, G. N., Nilsson, J. & Bengtsson, E. Autoantibodies against aldehyde-modified collagen type IV are associated with risk of development of myocardial infarction. J. Intern. Med. 282, 496–507 (2017).
Hudson, B. G., Tryggvason, K., Sundaramoorthy, M. & Neilson, E. G. Alport’s syndrome, Goodpasture’s syndrome, and type IV collagen. N. Engl. J. Med 348, 2543–2556 (2003).
Wang, Y. et al. COL4A3 gene variants and diabetic kidney disease in MODY. Clin. J. Am. Soc. Nephrol. 13, 1162–1171 (2018).
Acknowledgements
We acknowledge the use of cloud computing credits from the National Institutes of Health. M.M.R. was supported by a fellowship from the Deutsche Forschungsgemeinschaft (DFG; RI2811/1-1). This research was partially funded by US National Institutes of Health grants R35 GM130385 (G.S.), P30 MH062261 (G.S.), P01 DA026146 (G.S.) and U01 CA235493 (G.S.) and by Ecosystems and Networks Integrated with Genes and Molecular Assemblies (ENIGMA), a Scientific Focus Area Program at Lawrence Berkeley National Laboratory for the US Department of Energy, Office of Science, Office of Biological and Environmental Research, under contract number DE-AC02-05CH11231 (G.S.).
Author information
Authors and Affiliations
Contributions
E.L.-W.M. and E.M.B. led the protocol development and wrote the manuscript. H.P.B., A.P., C.G., M.M.R., X.D.-A. and J.R.M.-B. contributed ideas and data, tested the protocol and edited the manuscript. R.L.M. and B.A.T. assisted in protocol development and data analysis. R.S.P. and G.S. contributed ideas and edited the manuscript.
Corresponding author
Ethics declarations
Competing interests
Our initial interactions with IBM motivated these efforts; however, the technologies described herein are largely (>99%) independent of IBM.
Additional information
Peer review information Nature Protocols thanks Jianxin Chen, Alisdair Fernie and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key references using this protocol
Warth, B. et al. Anal. Chem. 89, 11505–11513 (2017): https://pubs.acs.org/doi/abs/10.1021/acs.analchem.7b02759
Rinschen, M. M. et al. Sci. Signal. 12, eaax9760 (2019): https://stke.sciencemag.org/content/12/611/eaax9760.abstract
Guijas, C. et al. Sci. Signal. 13, eabb2490 (2020): https://stke.sciencemag.org/content/13/648/eabb2490.full
Rinschen, M. M. et al. Nat. Rev. Mol. Cell Biol. 20, 353–367 (2019): https://www.nature.com/articles/s41580-019-0108-4
Domingo-Almenara, X. et al. Nat. Commun. 10, 5811 (2019): https://www.nature.com/articles/s41467-019-13680-7
Key data used in this protocol
Wikoff, W. R., Gangoiti, J. A., Barshop, B. A., & Siuzdak, G. Clin. Chem. 53, 2169–2176 (2007): https://academic.oup.com/clinchem/article/53/12/2169/5627367
Hyötyläinen, T. et al. Nat. Commun. 7, 8994 (2016): https://www.nature.com/articles/ncomms9994
Johnson, C. H. et al. Cell Metab. 21, 891–897 (2015): https://www.sciencedirect.com/science/article/pii/S1550413115001667
Huan, T. et al. Nat. Methods 14, 461–462 (2017): https://www.nature.com/articles/nmeth.4260
Supplementary information
Supplementary Information
Supplementary Procedures 1 and 2, Instructions for XCMS Systems Biology, Supplementary Tables 1–12 and Supplementary Figs. 1–14
Rights and permissions
About this article
Cite this article
Majumder, E.LW., Billings, E.M., Benton, H.P. et al. Cognitive analysis of metabolomics data for systems biology. Nat Protoc 16, 1376–1418 (2021). https://doi.org/10.1038/s41596-020-00455-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-020-00455-4
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.