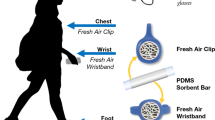
Abstract
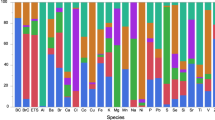
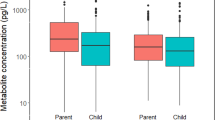
The complexity and dynamics of human diseases are driven by the interactions between internal molecular activities and external environmental exposures. Although advances in omics technology have dramatically broadened the understanding of internal molecular and cellular mechanisms, understanding of the external environmental exposures, especially at the personal level, is still rudimentary in comparison. This is largely owing to our limited ability to efficiently collect the personal environmental exposome (PEE) and extract the nucleic acids and chemicals from PEE. Here we describe a protocol that integrates hardware and experimental pipelines to collect and decode biotic and abiotic external exposome at the individual level. The described protocol has several advantages over conventional approaches, such as exposome monitoring at the personal level, decontamination steps to increase sensitivity and simultaneous capture and high-throughput profiling of biotic and abiotic exposures. The protocol takes ~18 h of bench time over 2–3 d to prepare samples for high-throughput profiling and up to a couple of weeks of instrumental time to analyze, depending on the number of samples. Hundreds to thousands of species and organic compounds could be detected in the airborne particulate samples using this protocol. The composition and complexity of the biotic and abiotic substances are heavily influenced by the sampling spatiotemporal factors. Basic skillsets in molecular biology and analytical chemistry are required to carry out this protocol. This protocol could be modified to decode biotic and abiotic substances in other types of low or ultra-low input samples.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The biotic exposome data generated using the protocol in the relevant study1 were deposited to the National Center of Biotechnology Information under bioproject ID PRJNA421162.
References
Jiang, C. et al. Dynamic human environmental exposome revealed by longitudinal personal monitoring. Cell 175, 277–291 (2018).
Huang, R. J. et al. High secondary aerosol contribution to particulate pollution during haze events in China. Nature 514, 218–222 (2015).
Cohen, S., Janicki-Deverts, D. & Miller, G. E. Psychological stress and disease. JAMA 298,1685–1687 (2007).
Vermeulen, R., Schymanski, E. L., Barabási, A. L. & Miller, G. W. The exposome and health: where chemistry meets biology. Science 367, 392–396 (2020).
Pfeifer, G. P. Environmental exposures and mutational patterns of cancer genomes. Genome Med. 2, 54 (2010).
Heederik, D. & Von Mutius, E. Does diversity of environmental microbial exposure matter for the occurrence of allergy and asthma? J. Allergy Clin. Immunol. 130, 44–50 (2012).
Miller, F. W. et al. Epidemiology of environmental exposures and human autoimmune diseases: findings from a National Institute of Environmental Health Sciences Expert Panel Workshop. J. Autoimmun. 39, 259–271 (2012).
Fujimura, K. E. et al. House dust exposure mediates gut microbiome Lactobacillus enrichment and airway immune defense against allergens and virus infection. Proc. Natl Acad. Sci. USA 111, 805–810 (2014).
Wild, C. P. Complementing the genome with an ‘exposome’: the outstanding challenge of environmental exposure measurement in molecular epidemiology. Cancer Epidemiol. Biomarkers Prev. 14, 1847–1850 (2005).
Wild, C. P. The exposome: from concept to utility. Int. J. Epidemiol. 41, 24–32 (2012).
Lindholm, M. E. et al. An integrative analysis reveals coordinated reprogramming of the epigenome and the transcriptome in human skeletal muscle after training. Epigenetics 9, 1557–1569 (2014).
Laker, R. C. et al. Transcriptomic and epigenetic responses to short-term nutrient-exercise stress in humans. Sci. Rep. 7, 15134 (2017).
Cao, C. et al. Inhalable microorganisms in Beijing’s PM2.5 and PM10 pollutants during a severe smog event. Environ. Sci. Technol. 48, 1499–1507 (2014).
Lax, S. et al. Longitudinal analysis of microbial interaction between humans and the indoor environment. Science 345, 1048–1052 (2014).
Dannemiller, K. C. et al. Next-generation DNA sequencing reveals that low fungal diversity in house dust is associated with childhood asthma development. Indoor Air 24, 236–247 (2014).
Barberán, A. et al. Continental-scale distributions of dust-associated bacteria and fungi. Proc. Natl Acad. Sci. USA 112, 5756–5761 (2015).
Abbatt, J. P. D. & Wang, C. The atmospheric chemistry of indoor environments. Environ. Sci. Process. Impacts 22, 25–48 (2020).
Oliveira, M., Slezakova, K., Delerue-Matos, C., Pereira, M. C. & Morais, S. Children environmental exposure to particulate matter and polycyclic aromatic hydrocarbons and biomonitoring in school environments: a review on indoor and outdoor exposure levels, major sources and health impacts. Environ Int. 124, 180–204 (2019).
Cissé, O. H., Ma, L., Jiang, C., Snyder, M. & Kovacs, J. A. Humans are selectively exposed to Pneumocystis jirovecii. mBio 11, e03138-19 (2020).
Hänninen, O. O. et al. The EXPOLIS study: implications for exposure research and environmental policy in Europe. J. Expo. Anal. Environ. Epidemiol. 14, 440–456 (2004).
Lelieveld, J., Evans, J. S., Fnais, M., Giannadaki, D. & Pozzer, A. The contribution of outdoor air pollution sources to premature mortality on a global scale. Nature 525, 367–371 (2015).
Behzad, H., Gojobori, T. & Mineta, K. Challenges and opportunities of airborne metagenomics. Genome Biol. Evol. 7, 1216–1226 (2015).
Jiang, W. et al. Optimized DNA extraction and metagenomic sequencing of airborne microbial communities. Nat. Protoc. 10, 768–779 (2015).
Gao, P., da Silva, E. B., Townsend, T., Liu, X. & Ma, L. Q. Emerging PAHs in urban soils: concentrations, bioaccessibility, and spatial distribution. Sci. Total Environ. 670, 800–805 (2019).
Xiang, P. et al. Cellular responses of normal (HL-7702) and cancerous (HepG2) hepatic cells to dust extract exposure. Chemosphere 193, 1189–1197 (2018).
Leung, M. H. Y., Wilkins, D., Li, E. K. T., Kong, F. K. F. & Lee, P. K. H. Indoor-air microbiome in an urban subway network: diversity and dynamics. Appl. Environ. Microbiol. 80, 6760–6770 (2014).
Hospodsky, D. et al. Characterizing airborne fungal and bacterial concentrations and emission rates in six occupied children’s classrooms. Indoor Air 25, 641–652 (2015).
Salter, S. J. et al. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol. 12, 87 (2014).
Weiss, S. et al. Tracking down the sources of experimental contamination in microbiome studies. Genome Biol. 15, 564 (2014).
Cui, Y. et al. The exposome: embracing the complexity for discovery in environmental health. Environ. Health Perspect. 124, A137–A140 (2016).
Gao, P. et al. Spatial and temporal changes of P and Ca distribution and fractionation in soil and sediment in a karst farmland-wetland system. Chemosphere 220, 844–650 (2019).
da Silva, E. B. et al. Background concentrations of trace metals As, Ba, Cd, Co, Cu, Ni, Pb, Se, and Zn in 214 Florida urban soils: different cities and land uses. Environ. Pollut. 264, 114737 (2020).
Volckens, J. et al. Development and evaluation of an ultrasonic personal aerosol sampler. Indoor Air 27, 409–416 (2017).
Contrepois, K. et al. Cross-platform comparison of untargeted and targeted lipidomics approaches on aging mouse plasma. Sci. Rep. 8, 17747 (2018).
Gao, P. et al. Human exposure to polycyclic aromatic hydrocarbons: metabolomics perspective. Environ. Int. 119, 466–477 (2018).
Gao, P. et al. Emerging and legacy PAHs in urban soils of four small cities: concentrations, distribution, and sources. Sci. Total Environ. 685, 463–470 (2019).
Wood, D. E., Lu, J. & Langmead, B. Improved metagenomic analysis with Kraken 2. Genome Biol. 20, 257 (2019).
Truong, D. T. et al. MetaPhlAn2 for enhanced metagenomic taxonomic profiling. Nat. Methods 12, 902–903 (2015).
Franzosa, E. A. et al. Species-level functional profiling of metagenomes and metatranscriptomes. Nat. Methods 15, 962–968 (2018).
Wishart, D. S. et al. HMDB 4.0: the human metabolome database for 2018. Nucleic Acids Res. 46, D608–D617 (2018).
Barupal, D. K. & Fiehn, O. Generating the blood exposome database using a comprehensive text mining and database fusion approach. Environ. Health Perspect. 127, 97008 (2019).
Wishart, D. et al. T3DB: the toxic exposome database. Nucleic Acids Res. 43, D928–D934 (2015).
García-Alcalde, F. et al. Qualimap: evaluating next-generation sequencing alignment data. Bioinformatics 28, 2678–2679 (2012).
Acknowledgements
We thank X. Wang and X. Li for their contributions to the development of the methods. This work was supported by Zhejiang University Life Sciences Institute start-up funds and the Leona M. and Harry B. Helmsley Charitable Trust (grant G-2004-03820).
Author information
Authors and Affiliations
Contributions
C.J. conceived the study. M.S. supervised the study. C.J., X.Z. and P.G. drafted and revised the protocol with input from Q.C. and M.S.
Corresponding authors
Ethics declarations
Competing interests
Two provisional patents were filed (pending application numbers 62/488119 and 62/488256).
Additional information
Peer review information Nature Protocols thanks Lea Maitre and Simone Morais for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key references using this protocol
Jiang C. et al. Cell 175, 277–291 (2018): https://doi.org/10.1016/j.cell.2018.08.060
Cissé O. et al. mBio 11, e03138-19 (2020): https://doi.org/10.1128/mBio.03138-19
Supplementary information
Supplementary Data 1
Cartridge design
Rights and permissions
About this article
Cite this article
Jiang, C., Zhang, X., Gao, P. et al. Decoding personal biotic and abiotic airborne exposome. Nat Protoc 16, 1129–1151 (2021). https://doi.org/10.1038/s41596-020-00451-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-020-00451-8
This article is cited by
-
A review on the application of the exposome paradigm to unveil the environmental determinants of age-related diseases
Human Genomics (2022)
-
Operationalizing the Exposome Using Passive Silicone Samplers
Current Pollution Reports (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.