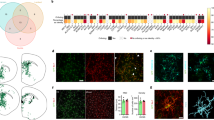
Abstract
Microglia are critically involved in complex neurological disorders with a strong genetic component, such as Alzheimer’s disease, Parkinson’s disease and frontotemporal dementia. Although mouse microglia can recapitulate aspects of human microglia physiology, they do not fully capture the human genetic aspects of disease and do not reproduce all human cell states. Primary cultures of human microglia or microglia derived from human induced pluripotent stem cells (PSCs) are difficult to maintain in brain-relevant cell states in vitro. Here we describe MIGRATE (microglia in vitro generation refined for advanced transplantation experiments, which provides a combined in vitro differentiation and in vivo xenotransplantation protocol to study human microglia in the context of the mouse brain. This article details an accurate, step-by-step workflow that includes in vitro microglia differentiation from human PSCs, transplantation into the mouse brain and quantitative analysis of engraftment. Compared to current differentiation and xenotransplantation protocols, we present an optimized, faster and more efficient approach that yields up to 80% chimerism. To quantitatively assess engraftment efficiency by flow cytometry, access to specialized flow cytometry is required. Alternatively, the percentage of chimerism can be estimated by standard immunohistochemical analysis. The MIGRATE protocol takes ~40 d to complete, from culturing PSCs to engraftment efficiency assessment.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Geirsdottir, L. et al. Cross-species single-cell analysis reveals divergence of the primate microglia program. Cell 179, 1609–1622 (2019).
Gosselin, D. et al. An environment-dependent transcriptional network specifies human microglia identity. Science 356, 1248–1259 (2017).
Masuda, T. et al. Spatial and temporal heterogeneity of mouse and human microglia at single-cell resolution. Nature 566, 388–392 (2019).
Mancuso, R. et al. Stem-cell-derived human microglia transplanted in mouse brain to study human disease. Nat. Neurosci. 22, 2111–2116 (2019).
Butovsky, O. et al. Identification of a unique TGF-β-dependent molecular and functional signature in microglia. Nat. Neurosci. 17, 131–143 (2014).
Muffat, J. et al. Efficient derivation of microglia-like cells from human pluripotent stem cells. Nat. Med. 22, 1358–1367 (2016).
Douvaras, P. et al. Directed differentiation of human pluripotent stem cells to microglia. Stem Cell Reports 8, 1516–1524 (2017).
Pandya, H. et al. Differentiation of human and murine induced pluripotent stem cells to microglia-like cells. Nat. Neurosci. 20, 753–759 (2017).
Abud, E. M. et al. iPSC-derived human microglia-like cells to study neurological diseases. Neuron 94, 278–293 (2017).
Haenseler, W. et al. A highly efficient human pluripotent stem cell microglia model displays a neuronal-co-culture-specific expression profile and inflammatory response. Stem Cell Reports 8, 1727–1742 (2017).
Takata, K. et al. Induced-pluripotent-stem-cell-derived primitive macrophages provide a platform for modeling tissue-resident macrophage differentiation and function. Immunity 47, 183–198 (2017).
Claes, C. et al. Human stem cell–derived monocytes and microglia-like cells reveal impaired amyloid plaque clearance upon heterozygous or homozygous loss of TREM2. Alzheimers Dement. 15, 453–464 (2019).
Timmerman, R., Burm, S. M. & Bajramovic, J. J. An overview of in vitro methods to study microglia. Front. Cell Neurosci. 12, 1–12 (2018).
Pocock, J. M. & Piers, T. M. Modelling microglial function with induced pluripotent stem cells: an update. Nat. Rev. Neurosci. 19, 445–452 (2018).
Haenseler, W. & Rajendran, L. Concise review: modeling neurodegenerative diseases with human pluripotent stem cell-derived microglia. Stem Cells 37, 724–730 (2019).
Bohlen, C. J. et al. Diverse requirements for microglial survival, specification, and function revealed by defined-medium cultures. Neuron 94, 759–773 (2017).
Hasselmann, J. et al. Development of a chimeric model to study and manipulate human microglia in vivo. Neuron 103, 1016–1033 (2019).
De Strooper, B. & Karran, E. The cellular phase of Alzheimer’s disease. Cell 164, 603–615 (2016).
Chen, W.-T. et al. Spatial transcriptomics and in situ sequencing to study Alzheimer’s disease. Cell 182, 976–991.e19 (2020).
Svoboda, D. S. et al. Human iPSC-derived microglia assume a primary microglia-like state after transplantation into the neonatal mouse brain. Proc. Natl Acad. Sci. USA 116, 25293–25303 (2019).
Xu, R. et al. Human iPSC-derived mature microglia retain their identity and functionally integrate in the chimeric mouse brain. Nat. Commun. 11, 1577 (2020).
McQuade, A. et al. Development and validation of a simplified method to generate human microglia from pluripotent stem cells. Mol. Neurodegener. 13, 1–13 (2018).
Yanagimachi, M. D. et al. Robust and highly-efficient differentiation of functional monocytic cells from human pluripotent stem cells under serum- and feeder cell-free conditions. PLoS ONE 8, 1–9 (2013).
van Wilgenburg, B., Browne, C., Vowles, J. & Cowley, S. A. Efficient, long term production of monocyte-derived macrophages from human pluripotent stem cells under partly-defined and fully-defined conditions. PLoS ONE 8, e71098 (2013).
Anderson, J. et al. Derivation of normal macrophages from human embryonic stem (hES) cells for applications in HIV gene therapy. Retrovirology 3, 24 (2006).
Subramanian, A. et al. Macrophage differentiation from embryoid bodies derived from human embryonic stem cells. J. Stem Cells 4, 29–45 (2009).
Choi, K. D., Vodyanik, M. & Slukvin, I. I. Hematopoietic differentiation and production of mature myeloid cells from human pluripotent stem cells. Nat. Protoc. 6, 296–313 (2011).
Rathinam, C. et al. Efficient differentiation and function of human macrophages in humanized CSF-1 mice. Blood 118, 3119–3128 (2011).
Pick, M., Azzola, L., Mossman, A., Stanley, E. G. & Elefanty, A. G. Differentiation of human embryonic stem cells in serum-free medium reveals distinct roles for bone morphogenetic protein 4, vascular endothelial growth factor, stem cell factor, and fibroblast growth factor 2 in hematopoiesis. Stem Cells 25, 2206–2214 (2007).
Mathys, H. et al. Single-cell transcriptomic analysis of Alzheimer’s disease. Nature 570, 332–337 (2019).
Lambert, J. C. et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat. Genet. 45, 1452–1458 (2013).
Kunkle, B. W. et al. Genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates Aβ, tau, immunity and lipid processing. Nat. Genet. 51, 414–430 (2019).
Jansen, I. E. et al. Genome-wide meta-analysis identifies new loci and functional pathways influencing Alzheimer’s disease risk. Nat. Genet. 51, 404–413 (2019).
Paloneva, J., Autti, T., Hakola, P. & Haltia, M. J. Polycystic lipomembranous osteodysplasia with sclerosing leukoencephalopathy (PLOSL). GeneReviews https://www.ncbi.nlm.nih.gov/books/NBK1197 (2002).
Keo, A. et al. Transcriptomic signatures of brain regional vulnerability to Parkinson’s disease. Commun. Biol 3, 1–12 (2020).
Wang, L. et al. CD200 maintains the region-specific phenotype of microglia in the midbrain and its role in Parkinson’s disease. Glia 68, 1874–1890 (2020).
Haenseler, W. et al. Excess α-synuclein compromises phagocytosis in iPSC-derived macrophages. Sci. Rep. 7, 1–11 (2017).
Dwyer, Z. et al. Leucine-rich repeat kinase-2 (LRRK2) modulates microglial phenotype and dopaminergic neurodegeneration. Neurobiol. Aging 91, 45–55 (2020).
Kuter, K. Z., Cenci, M. A. & Carta, A. R. The role of glia in Parkinson’s disease: emerging concepts and therapeutic applications. Prog. Brain Res. 252, 131–168 (2020).
Hong, S. et al. Complement and microglia mediate early synapse loss in Alzheimer mouse models. Science 352, 1–9 (2016).
Sekar, A. et al. Schizophrenia risk from complex variation of complement component 4. Nature 530, 177–183 (2016).
Sellgren, C. M. et al. Increased synapse elimination by microglia in schizophrenia patient-derived models of synaptic pruning. Nat. Neurosci. 22, 374–385 (2019).
Swanstrom, R. & Coffin, J. HIV-1 pathogenesis: the virus. Cold Spring Harb. Perspect. Med. 2, a007443 (2012).
Spudich, S. et al. HIV-1-related central nervous system disease: current issues in pathogenesis, diagnosis, and treatment. Cold Spring Harb. Perspect. Med. 2, a007120 (2012).
Uncini, A., Shahrizaila, N. & Kuwabara, S. Zika virus infection and Guillain–Barré syndrome: a review focused on clinical and electrophysiological subtypes. J. Neurol. Neurosurg. Psychiatry 88, 266–271 (2017).
Martí, M. et al. Characterization of pluripotent stem cells. Nat. Protoc. 8, 223–253 (2013).
Acknowledgements
This project received funding from the European Research Council (ERC) under the European Union’s Horizon 2020 Research and Innovation Programme (grant agreement no. ERC-834682 CELLPHASE_AD). This work was also supported by UK Dementia Research, the Flanders Institute for Biotechnology (VIB vzw), Vlaams Initiatief voor Netwerken voor Dementie Onderzoek (Strategic Basic Research Grant no. 135043), a Methusalem grant from KU Leuven and the Flemish Government, the Fonds voor Wetenschappelijk Onderzoek, KU Leuven, The Queen Elisabeth Medical Foundation for Neurosciences, the Opening the Future campaign of the Leuven Universitair Fonds, the Belgian Alzheimer Research Foundation and the Alzheimer’s Association USA. R.M. has funding from Fonds voor Wetenschappelijk Onderzoek (grant no. G0C9219N) and is a recipient of a postdoctoral fellowship from the Alzheimer’s Association USA (fellowship no. 2018-AARF-591110). B.D.S. holds the Bax-Vanluffelen Chair for Alzheimer’s Disease. B.D.S. receives funding from the Medical Research Council, the Alzheimer’s Society and Alzheimer’s Research UK via the Dementia Research Institute. N.F. is recipient of a PhD fellowship from Fonds voor Wetenschappelijk Onderzoek (fellowship no. 1139520N). A.M.-M. is supported by a fellowship from the Alzheimer’s Association USA (fellowship no. AARF-20-684397) and a Marie Skłodowska-Curie Actions - Seal of Excellence Postdoctoral Fellowship. We thank V. Hendrickx and J. Verwaeren for animal husbandry. Imaging was acquired through Nikon A1R Eclipse Ti confocal from LiMoNe facility at CBD. We thank S. Balusu and J. Van Den Daele for providing stem cell expertise.
Author information
Authors and Affiliations
Contributions
N.F., A.M.-M., B.D.S. and R.M. conceived the study. N.F., A.M.-M. and R.M. performed all the experiments and wrote the manuscript. I.G. and L.W. assisted with microglia xenotransplantations and engraftment efficiency analysis. All authors read and approved the final manuscript for publication.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests. B.D.S. receives grants from different companies that support his research and is a consultant for several companies, but nothing is directly related to the current publication.
Additional information
Peer review information Nature Protocols thanks Valentina Fossati and Abed Alfattah Mansour for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key reference using this protocol
Mancuso, R. et al. Nat. Neurosci. 22, 2111–2116 (2019): https://doi.org/10.1038/s41593-019-0525-x
Extended data
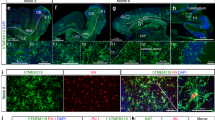
Extended Data Fig. 1 MIGRATE protocol representative pictures.
Representative day-by-day pictures of the MIGRATE differentiation protocol (H9 cells). The plates used at the two main stages are reported: days 0 to 4 are performed in low-adherent U-form 96-well plates; days 4 to 18 are performed in regular six-well plates. Media changes are indicated in the appropriate pictures with cytokines acronyms (see ‘Procedure’). Length of scale bars is indicated in the images (10 μm for days 15, 17 and 18; 100 μm for all other pictures).
Supplementary information
Supplementary Information
Supplementary Methods (histological analysis), Supplementary Tables 1 and 2 and Supplementary Fig. 1.
Rights and permissions
About this article
Cite this article
Fattorelli, N., Martinez-Muriana, A., Wolfs, L. et al. Stem-cell-derived human microglia transplanted into mouse brain to study human disease. Nat Protoc 16, 1013–1033 (2021). https://doi.org/10.1038/s41596-020-00447-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-020-00447-4
This article is cited by
-
Regulation of human microglial gene expression and function via RNAase-H active antisense oligonucleotides in vivo in Alzheimer’s disease
Molecular Neurodegeneration (2024)
-
Impact of microglia isolation and culture methodology on transcriptional profile and function
Journal of Neuroinflammation (2024)
-
Xenografted human microglia display diverse transcriptomic states in response to Alzheimer’s disease-related amyloid-β pathology
Nature Neuroscience (2024)
-
Crosstalk Among Glial Cells in the Blood–Brain Barrier Injury After Ischemic Stroke
Molecular Neurobiology (2024)
-
Alzheimer’s genes in microglia: a risk worth investigating
Molecular Neurodegeneration (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.