Abstract
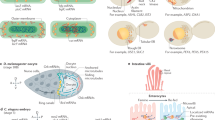
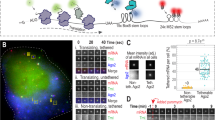
mRNA translation is a key step in gene expression. Proper regulation of translation efficiency ensures correct protein expression levels in the cell, which is essential to cell function. Different methods used to study translational control in the cell rely on population-based assays that do not provide information about translational heterogeneity between cells or between mRNAs of the same gene within a cell, and generally provide only a snapshot of translation. To study translational heterogeneity and measure translation dynamics, we have developed microscopy-based methods that enable visualization of translation of single mRNAs in live cells. These methods consist of a set of genetic tools, an imaging-based approach and sophisticated computational tools. Using the translation imaging method, one can investigate many new aspects of translation in single living cells, such as translation start-site selection, 3ʹ-UTR (untranslated region) translation and translation-coupled mRNA decay. Here, we describe in detail how to perform such experiments, including reporter design, cell line generation, image acquisition and analysis. This protocol also provides a detailed description of the image analysis pipeline and computational modeling that will enable non-experts to correctly interpret fluorescence measurements. The protocol takes 2–4 d to complete (after cell lines expressing all required transgenes have been generated).
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Code availability
The analysis software (RiboFitter and TransTrack) is included in Supplementary Data 1 and 2, and is also freely available through GitHub (http://github.com/TanenbaumLab/RiboFitter and http://github.com/TanenbaumLab/TransTrack, respectively). The code in this manuscript has been peer-reviewed.
References
Tanenbaum, M. E., Stern-Ginossar, N., Weissman, J. S. & Vale, R. D. Regulation of mRNA translation during mitosis. eLife 4, e07957 (2015).
Trcek, T., Larson, D. R., Moldon, A., Query, C. C. & Singer, R. H. Single-molecule mRNA decay measurements reveal promoter- regulated mRNA stability in yeast. Cell 147, 1484–1497 (2011).
Ephrussi, A. & Lehmann, R. Induction of germ cell formation by oskar. Nature 358, 387–392 (1992).
Lasko, P. mRNA localization and translational control in Drosophila oogenesis. Cold Spring Harb. Perspect. Biol. 4, a012294 (2012).
Holt, C. E. & Schuman, E. M. The central dogma decentralized: new perspectives on RNA function and local translation in neurons. Neuron 80, 648–657 (2013).
Shoemaker, C. J. & Green, R. Translation drives mRNA quality control. Nat. Struct. Mol. Biol. 19, 594–601 (2012).
Schwanhausser, B. et al. Global quantification of mammalian gene expression control. Nature 473, 337–342 (2011).
Pelechano, V., Wei, W. & Steinmetz, L. M. Widespread co-translational RNA decay reveals ribosome dynamics. Cell 161, 1400–1412 (2015).
Hu, W., Sweet, T. J., Chamnongpol, S., Baker, K. E. & Coller, J. Co-translational mRNA decay in Saccharomyces cerevisiae. Nature 461, 225–229 (2009).
He, F. & Jacobson, A. Nonsense-mediated mRNA decay: degradation of defective transcripts is only part of the story. Annu. Rev. Genet. 49, 339–366 (2015).
Yan, X., Hoek, T. A., Vale, R. D. & Tanenbaum, M. E. Dynamics of translation of single mRNA molecules in vivo. Cell 165, 976–989 (2016).
Ruijtenberg, S., Hoek, T. A., Yan, X. & Tanenbaum, M. E. Imaging translation dynamics of single mRNA molecules in live cells. Methods Mol. Biol. 1649, 385–404 (2018).
Morisaki, T. et al. Real-time quantification of single RNA translation dynamics in living cells. Science 352, 1425–1429 (2016).
Wang, C., Han, B., Zhou, R. & Zhuang, X. Real-time imaging of translation on single mRNA transcripts in live cells. Cell 165, 990–1001 (2016).
Wu, B., Eliscovich, C., Yoon, Y. J. & Singer, R. H. Translation dynamics of single mRNAs in live cells and neurons. Science 352, 1430–1435 (2016).
Pichon, X. et al. Visualization of single endogenous polysomes reveals the dynamics of translation in live human cells. J. Cell Biol. 214, 769–781 (2016).
Tanenbaum, M. E., Gilbert, L. A., Qi, L. S., Weissman, J. S. & Vale, R. D. A protein-tagging system for signal amplification in gene expression and fluorescence imaging. Cell 159, 635–646 (2014).
Chao, J. A., Patskovsky, Y., Almo, S. C. & Singer, R. H. Structural basis for the coevolution of a viral RNA-protein complex. Nat. Struct. Mol. Biol. 15, 103–105 (2008).
Atkins, J. F., Loughran, G., Bhatt, P. R., Firth, A. E. & Baranov, P. V. Ribosomal frameshifting and transcriptional slippage: From genetic steganography and cryptography to adventitious use. Nucleic Acids Res. 44, 7007–7078 (2016).
Calvo, S. E., Pagliarini, D. J. & Mootha, V. K. Upstream open reading frames cause widespread reduction of protein expression and are polymorphic among humans. Proc. Natl Acad. Sci. USA 106, 7507–7512 (2009).
Boersma, S. et al. Multi-color single-molecule imaging uncovers extensive heterogeneity in mRNA decoding. Cell 178, 458–472.e419 (2019).
Lyon, K. et al. Live-cell single RNA imaging reveals bursts of translational frameshifting. Mol. Cell 75, 172–183.e9 (2019).
Hoek, T. A. et al. Single-molecule imaging uncovers rules governing nonsense-mediated mRNA decay. Mol. Cell 75, 324-339.e11 (2019).
He, F. et al. Genome-wide analysis of mRNAs regulated by the nonsense-mediated and 5ʹ to 3ʹ mRNA decay pathways in yeast. Mol. Cell 12, 1439–1452 (2003).
Ingolia, N. T., Ghaemmaghami, S., Newman, J. R. & Weissman, J. S. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324, 218–223 (2009).
Ingolia, N. T., Lareau, L. F. & Weissman, J. S. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 147, 789–802 (2011).
Brittis, P. A., Lu, Q. & Flanagan, J. G. Axonal protein synthesis provides a mechanism for localized regulation at an intermediate target. Cell 110, 223–235 (2002).
Aakalu, G., Smith, W. B., Nguyen, N., Jiang, C. & Schuman, E. M. Dynamic visualization of local protein synthesis in hippocampal neurons. Neuron 30, 489–502 (2001).
Han, K. et al. Parallel measurement of dynamic changes in translation rates in single cells. Nat. Methods 11, 86–93 (2014).
Leung, K. M. et al. Asymmetrical β-actin mRNA translation in growth cones mediates attractive turning to netrin-1. Nat. Neurosci. 9, 1247–1256 (2006).
Yu, J., Xiao, J., Ren, X., Lao, K. & Xie, X. S. Probing gene expression in live cells, one protein molecule at a time. Science 311, 1600–1603 (2006).
Rodriguez, A. J., Shenoy, S. M., Singer, R. H. & Condeelis, J. Visualization of mRNA translation in living cells. J. Cell Biol. 175, 67–76 (2006).
Halstead, J. M. et al. Translation. An RNA biosensor for imaging the first round of translation from single cells to living animals. Science 347, 1367–1671 (2015).
Katz, Z. B. et al. Mapping translation ‘hot-spots’ in live cells by tracking single molecules of mRNA and ribosomes. eLife 5, e10415 (2016).
Wu, B., Buxbaum, A. R., Katz, Z. B., Yoon, Y. J. & Singer, R. H. Quantifying protein-mRNA interactions in single live cells. Cell 162, 211–220 (2015).
Bertrand, E. et al. Localization of ASH1 mRNA particles in living yeast. Mol. Cell 2, 437–445 (1998).
Dunn, J. G., Foo, C. K., Belletier, N. G., Gavis, E. R. & Weissman, J. S. Ribosome profiling reveals pervasive and regulated stop codon readthrough in Drosophila melanogaster. eLife 2, e01179 (2013).
Young, D. J., Guydosh, N. R., Zhang, F., Hinnebusch, A. G. & Green, R. Rli1/ABCE1 recycles terminating ribosomes and controls translation reinitiation in 3ʹUTRs in vivo. Cell 162, 872–884 (2015).
Gossen, M. et al. Transcriptional activation by tetracyclines in mammalian cells. Science 268, 1766–1769 (1995).
Edelstein, A., Amodaj, N., Hoover, K., Vale, R. & Stuurman, N. Computer control of microscopes using µManager. Curr. Protoc. Mol. Biol., 92, 14.20 (2010).
Grimm, J. B. et al. A general method to improve fluorophores for live-cell and single-molecule microscopy. Nat. Methods 12, 244–250 (2015).
Acknowledgements
We thank the members of the Tanenbaum lab for helpful discussions. This work was financially supported by an ERC starting grant (ERC-STG 677936-RNAREG), two grants from the Netherlands Organization for Scientific Research (NWO; ALWOP.290 and NWO/016.VIDI.189.005), and the Howard Hughes Medical Institute through an International Research Scholar grant to M.E.T. (HHMI/IRS 55008747). All authors were supported by the Oncode Institute, which is partly funded by the Dutch Cancer Society (KWF). T.A.H. was supported by a PhD fellowship from the Boehringer Ingelheim Fonds.
Author information
Authors and Affiliations
Contributions
D.K., T.A.H. and S.S. made the figures in the paper. D.K., T.A.H. and M.E.T. wrote the manuscript with input from S.S., B.M.P.V. and S.B.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Protocols thanks Niels Gehring and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key references using this protocol
Boersma, S. et al. Cell 178, 458–472.e419 (2019): https://doi.org/10.1016/j.cell.2019.05.001
Hoek, T. et al. Mol. Cell 75, 324–339.e11 (2019): https://doi.org/10.1016/j.molcel.2019.05.008
Supplementary information
Supplementary Data 1
TransTrack software, example data, and a tutorial on how to use TransTrack.
Supplementary Data 2
RiboFitter software, example data, and expected outcomes of RiboFitter analysis.
Supplementary Data 3
Excel template for making a Kaplan–Meier plot.
Rights and permissions
About this article
Cite this article
Khuperkar, D., Hoek, T.A., Sonneveld, S. et al. Quantification of mRNA translation in live cells using single-molecule imaging. Nat Protoc 15, 1371–1398 (2020). https://doi.org/10.1038/s41596-019-0284-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-019-0284-x
This article is cited by
-
Single-cell measurement of plasmid copy number and promoter activity
Nature Communications (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.