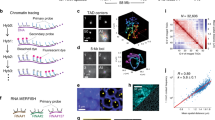
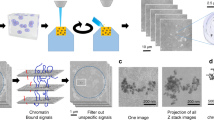
Abstract
During DNA replication, the genetic information of a cell is copied. Subsequently, identical genetic information is segregated reliably to the two daughter cells through cell division. Meanwhile, DNA replication is intrinsically linked to the process of chromatin duplication, which is required for regulating gene expression and establishing cell identities. Understanding how chromatin is established, maintained or changed during DNA replication represents a fundamental question in biology. Recently, we developed a method to directly visualize chromatin components at individual replication forks undergoing DNA replication. This method builds upon the existing chromatin fiber technique and combines it with cell type–specific chromatin labeling and superresolution microscopy. In this method, a short pulse of nucleoside analog labels replicative regions in the cells of interest. Chromatin fibers are subsequently isolated and attached to a glass slide, after which a laminar flow of lysis buffer extends the lysed chromatin fibers parallel with the direction of the flow. Fibers are then immunostained for different chromatin-associated proteins and mounted for visualization using superresolution microscopy. Replication foci, or ‘bubbles,’ are identified by the presence of the incorporated nucleoside analog. For researchers experienced in molecular biology and superresolution microscopy, this protocol typically takes 2–3 d from sample preparation to data acquisition, with an additional day for data processing and quantification.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Kornberg, R. D. & Lorch, Y. Chromatin structure and transcription. Annu. Rev. Cell Biol. 8, 563–587 (1992).
Snedeker, J., Wooten, M. & Chen, X. The inherent asymmetry of DNA replication. Annu. Rev. Cell Dev. Biol. 33, 291–318 (2017).
Burgers, P. M. J. & Kunkel, T. A. Eukaryotic DNA replication fork. Annu. Rev. Biochem. 86, 417–438 (2017).
Bell, S. P. & Dutta, A. DNA replication in eukaryotic cells. Annu. Rev. Biochem. 71, 333–374 (2002).
DePamphilis, M. L. Review: nuclear structure and DNA replication. J. Struct. Biol. 129, 186–197 (2000).
Alabert, C., Jasencakova, Z. & Groth, A. Chromatin replication and histone dynamics. Adv. Exp. Med. Biol. 1042, 311–333 (2017).
Ramachandran, S., Ahmad, K. & Henikoff, S. Capitalizing on disaster: establishing chromatin specificity behind the replication fork. Bioessays 39 (2017).
Miller, T. C. & Costa, A. The architecture and function of the chromatin replication machinery. Curr. Opin. Struct. Biol. 47, 9–16 (2017).
Ehrenhofer-Murray, A. E., Kamakaka, R. T. & Rine, J. A role for the replication proteins PCNA, RF-C, polymerase epsilon and Cdc45 in transcriptional silencing in Saccharomyces cerevisiae. Genetics 153, 1171–1182 (1999).
Luger, K., Mader, A. W., Richmond, R. K., Sargent, D. F. & Richmond, T. J. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 389, 251–260 (1997).
Arents, G., Burlingame, R. W., Wang, B. C., Love, W. E. & Moudrianakis, E. N. The nucleosomal core histone octamer at 3.1 Å resolution: a tripartite protein assembly and a left-handed superhelix. Proc. Natl Acad. Sci. USA 88, 10148–10152 (1991).
Annunziato, A. T. Assembling chromatin: the long and winding road. Biochim. Biophys. Acta 1819, 196–210 (2013).
Hammond, C. M., Stromme, C. B., Huang, H., Patel, D. J. & Groth, A. Histone chaperone networks shaping chromatin function. Nat. Rev. Mol. Cell. Biol. 18, 141–158 (2017).
Worcel, A., Han, S. & Wong, M. L. Assembly of newly replicated chromatin. Cell 15, 969–977 (1978).
Sogo, J. M., Stahl, H., Koller, T. & Knippers, R. Structure of replicating simian virus 40 minichromosomes. The replication fork, core histone segregation and terminal structures. J. Mol. Biol. 189, 189–204 (1986).
Jackson, V. & Chalkley, R. A new method for the isolation of replicative chromatin: selective deposition of histone on both new and old DNA. Cell 23, 121–134 (1981).
Leffak, I. M., Grainger, R. & Weintraub, H. Conservative assembly and segregation of nucleosomal histones. Cell 12, 837–845 (1977).
Seidman, M. M., Levine, A. J. & Weintraub, H. The asymmetric segregation of parental nucleosomes during chrosome replication. Cell 18, 439–449 (1979).
Weintraub, H. Cooperative alignment of nu bodies during chromosome replication in the presence of cycloheximide. Cell 9, 419–422 (1976).
Roufa, D. J. & Marchionni, M. A. Nucleosome segregation at a defined mammalian chromosomal site. Proc. Natl Acad. Sci. USA 79, 1810–1814 (1982).
Yu, C. et al. A mechanism for preventing asymmetric histone segregation onto replicating DNA strands. Science 361, 1386–1389 (2018).
Petryk, N. et al. MCM2 promotes symmetric inheritance of modified histones during DNA replication. Science 361, 1389–1392 (2018).
Wooten, M. et al. Asymmetric histone inheritance via strand-specific incorporation and biased replication fork movement. Nat. Struct. Mol. Biol. 26, 732–743 (2019).
Alabert, C. et al. Two distinct modes for propagation of histone PTMs across the cell cycle. Genes Dev. 29, 585–590 (2015).
Lin, S., Yuan, Z. F., Han, Y., Marchione, D. M. & Garcia, B. A. Preferential phosphorylation on old histones during early mitosis in human cells. J. Biol. Chem. 291, 15342–15357 (2016).
Yu, C. et al. Strand-specific analysis shows protein binding at replication forks and PCNA unloading from lagging strands when forks stall. Mol. Cell 56, 551–563 (2014).
Petryk, N. et al. Replication landscape of the human genome. Nat. Commun. 7, 10208 (2016).
McKnight, S. L. & Miller, O. L. Jr. Electron microscopic analysis of chromatin replication in the cellular blastoderm Drosophila melanogaster embryo. Cell 12, 795–804 (1977).
Lafzi, A., Moutinho, C., Picelli, S. & Heyn, H. Tutorial: guidelines for the experimental design of single-cell RNA sequencing studies. Nat. Protoc. 13, 2742–2757 (2018).
Chen, G., Ning, B. & Shi, T. Single-cell RNA-seq technologies and related computational data analysis. Front. Genet. 10, 317 (2019).
Cohen, S. M., Chastain, P. D. 2nd, Cordeiro-Stone, M. & Kaufman, D. G. DNA replication and the GINS complex: localization on extended chromatin fibers. Epigenetics Chromatin 2, 6 (2009).
Ahmad, K. & Henikoff, S. Histone H3 variants specify modes of chromatin assembly. Proc. Natl Acad. Sci. USA 99(Suppl 4), 16477–16484 (2002).
Blower, M. D., Sullivan, B. A. & Karpen, G. H. Conserved organization of centromeric chromatin in flies and humans. Dev. Cell 2, 319–330 (2002).
Chang, C. H. et al. Islands of retroelements are major components of Drosophila centromeres. PLOS Biol. 17, e3000241 (2019).
Kuzminov, A. When DNA topology turns deadly—RNA polymerases dig in their R-Loops to stand their ground: new positive and negative (super)twists in the replication-transcription conflict. Trends Genet. 34, 111–120 (2018).
Koster, D. A., Crut, A., Shuman, S., Bjornsti, M. A. & Dekker, N. H. Cellular strategies for regulating DNA supercoiling: a single-molecule perspective. Cell 142, 519–530 (2010).
Wang, J. C. Cellular roles of DNA topoisomerases: a molecular perspective. Nat. Rev. Mol. Cell. Biol. 3, 430–440 (2002).
Ljungman, M. & Hanawalt, P. C. Localized torsional tension in the DNA of human cells. Proc. Natl Acad. Sci. USA 89, 6055–6059 (1992).
LaMarr, W. A., Yu, L., Nicolaou, K. C. & Dedon, P. C. Supercoiling affects the accessibility of glutathione to DNA-bound molecules: positive supercoiling inhibits calicheamicin-induced DNA damage. Proc. Natl Acad. Sci. USA 95, 102–107 (1998).
Hell, S. W. & Wichmann, J. Breaking the diffraction resolution limit by stimulated emission: stimulated-emission-depletion fluorescence microscopy. Opt. Lett. 19, 780–782 (1994).
Sivaguru, M. et al. Comparative performance of airyscan and structured illumination superresolution microscopy in the study of the surface texture and 3D shape of pollen. Microsc. Res. Tech. 81, 101–114 (2018).
Gustafsson, M. G. Surpassing the lateral resolution limit by a factor of two using structured illumination microscopy. J. Microsc. 198, 82–87 (2000).
Betzig, E. et al. Imaging intracellular fluorescent proteins at nanometer resolution. Science 313, 1642–1645 (2006).
Rust, M. J., Bates, M. & Zhuang, X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nat. Methods 3, 793–795 (2006).
Glushonkov, O., Réal, E., Boutant, E., Mély, Y. & Didier, P. Optimized protocol for combined PALM-dSTORM imaging. Sci. Rep. 8, 8749 (2018).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Techer, H. et al. Replication dynamics: biases and robustness of DNA fiber analysis. J. Mol. Biol. 425, 4845–4855 (2013).
Blythe, S. A. & Wieschaus, E. F. Establishment and maintenance of heritable chromatin structure during early Drosophila embryogenesis. elife 5, e20148 (2016).
White-Cooper, H. Spermatogenesis: analysis of meiosis and morphogenesis. Methods Mol. Biol. 247, 45–75 (2004).
Huang, H. et al. A unique binding mode enables MCM2 to chaperone histones H3-H4 at replication forks. Nat. Struct. Mol. Biol. 22, 618–626 (2015).
Gan, H. et al. The Mcm2-Ctf4-Polalpha axis facilitates parental histone H3-H4 transfer to lagging strands. Mol. Cell 72, 140–151.e3 (2018).
Gan, H. et al. Checkpoint kinase Rad53 couples leading- and lagging-strand DNA synthesis under replication stress. Mol. Cell 68, 446–455.e3 (2017).
Zhou, J. C. et al. CMG-Pol epsilon dynamics suggests a mechanism for the establishment of leading-strand synthesis in the eukaryotic replisome. Proc. Natl Acad. Sci. USA 114, 4141–4146 (2017).
Acknowledgements
We thank Chuanhe Yu, Xu Hua and Zhiguo Zhang as well as Nataliya Petryk and Anja Groth for helpful discussions of this manuscript. We thank Shelby Blythe and Eric Wieschaus for the PCNA-EGFP fly line. We thank Barbara Mellone and Sharon Pavanacherry for suggestions on the chromatin fiber technique. We thank Johns Hopkins Integrated Imaging Center for confocal and Airyscan imaging and Carnegie Institute Imaging Center for STED microscopy work. This work was supported by NIH grants 5T32GM007231 and F31GM115149-01A1 (M.W.), NIH grant T32GM007231 (J.S.), NIH grant R01GM33397 (J.G.G.), and NIH grants R35GM127075 and R01GM112008, the Howard Hughes Medical Institute, the David and Lucile Packard Foundation and Johns Hopkins University startup funds (X.C.)
Author information
Authors and Affiliations
Contributions
Conceptualization, M.W., J.S., Z.F.N., J.G.G. and X.C.; methodology, M.W., J.S., Y.L., Z.F.N., J.G.G. and X.C.; investigation, M.W. and Y.L.; writing original draft, M.W. and X.C.; funding acquisition, J.G.G. and X.C.; supervision, J.G.G. and X.C.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Protocols thanks Robert Duronio, Bing Zhu and Corella Casas-Delucchi for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key reference using this protocol
Wooten, M. et al. Nat. Struct. Mol. Biol. 26, 732–743 (2019): https://doi.org/10.1038/s41594-019-0269-z
Integrated supplementary information
Supplementary Fig. 1 Modified 50-ml conical tube.
A modified 50-ml conical tube with a small hole made at the bottom with a 25-gauge hypodermic needle.
Supplementary Fig. 2 Modified 50-ml conical tube with lysis buffer.
Modified 50-ml conical tube containing 25 ml of lysis buffer with the cap screwed on and angled at 35° in preparation for lysis and fiber-generation steps.
Supplementary Fig. 3 50-ml conical tube zoomed-in view.
A zoom-in picture of Supplementary Fig. 2.
Supplementary Fig. 4 Humid chamber closed.
A humid chamber made with a P1000 tip box. The lid is wrapped with tin foil to minimize light exposure to slides placed inside. The lid should be placed on when slides are inside. The humid chamber can contain four slides comfortably.
Supplementary Fig. 5 Humid chamber open.
A humid chamber with the lid off. Note that a wet paper towel is placed at the bottom of the tip box shortly before adding the slides, which are placed on top of the tip rack.
Supplementary information
Supplementary Information
Supplementary Figs. 1–5, Supplementary Tables 1 and 2 and Supplementary Methods.
Rights and permissions
About this article
Cite this article
Wooten, M., Li, Y., Snedeker, J. et al. Superresolution imaging of chromatin fibers to visualize epigenetic information on replicative DNA. Nat Protoc 15, 1188–1208 (2020). https://doi.org/10.1038/s41596-019-0283-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-019-0283-y
This article is cited by
-
Dynamic de novo heterochromatin assembly and disassembly at replication forks ensures fork stability
Nature Cell Biology (2023)
-
Efficient and strand-specific profiling of replicating chromatin with enrichment and sequencing of protein-associated nascent DNA in mammalian cells
Nature Protocols (2021)
-
A latent subset of human hematopoietic stem cells resists regenerative stress to preserve stemness
Nature Immunology (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.