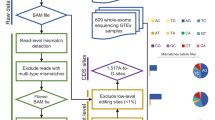
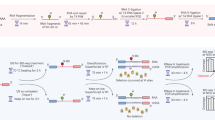
Abstract
RNA editing is a widespread post-transcriptional mechanism able to modify transcripts through insertions/deletions or base substitutions. It is prominent in mammals, in which millions of adenosines are deaminated to inosines by members of the ADAR family of enzymes. A-to-I RNA editing has a plethora of biological functions, but its detection in large-scale transcriptome datasets is still an unsolved computational task. To this aim, we developed REDItools, the first software package devoted to the RNA editing profiling in RNA-sequencing (RNAseq) data. It has been successfully used in human transcriptomes, proving the tissue and cell type specificity of RNA editing as well as its pervasive nature. Outcomes from large-scale REDItools analyses on human RNAseq data have been collected in our specialized REDIportal database, containing more than 4.5 million events. Here we describe in detail two bioinformatic procedures based on our computational resources, REDItools and REDIportal. In the first procedure, we outline a workflow to detect RNA editing in the human cell line NA12878, for which transcriptome and whole genome data are available. In the second procedure, we show how to identify dysregulated editing at specific recoding sites in post-mortem brain samples of Huntington disease donors. On a 64-bit computer running Linux with ≥32 GB of random-access memory (RAM), both procedures should take ~76 h, using 4 to 24 cores. Our protocols have been designed to investigate RNA editing in different organisms with available transcriptomic and/or genomic reads. Scripts to complete both procedures and a docker image are available at https://github.com/BioinfoUNIBA/REDItools.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Example datasets that include NA12878 RNAseq/WGS reads and RNAseq reads from BioProject PRJNA316625 are freely available at the ENA database (https://www.ebi.ac.uk/ena) or the SRA archive (https://www.ncbi.nlm.nih.gov/sra).
Code availability
REDItools source code is available at the GitHub website (https://github.com/BioinfoUNIBA/REDItools) under the MIT License. The code in this protocol has been peer reviewed.
References
Saletore, Y., Meyer, K., Korlach, J., Vilfan, I. D., Jaffrey, S. & Mason, C. E. The birth of the Epitranscriptome: deciphering the function of RNA modifications. Genome Biol. 13, 175 (2012).
Boccaletto, P. et al. MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res. 46, D303–D307 (2018).
Jantsch, M. F. & Schaefer, M. R. Mining the epitranscriptome: detection of RNA editing and RNA modifications. Methods 156, 1–4 (2019).
Benne, R. et al. Major transcript of the frameshifted coxII gene from trypanosome mitochondria contains four nucleotides that are not encoded in the DNA. Cell 46, 819–826 (1986).
Gott, J. M. & Emeson, R. B. Functions and mechanisms of RNA editing. Annu. Rev. Genet. 34, 499–531 (2000).
Eisenberg, E. & Levanon, E. Y. A-to-I RNA editing—immune protector and transcriptome diversifier. Nat. Rev. Genet. 19, 473–490 (2018).
Rosenberg, B. R., Hamilton, C. E., Mwangi, M. M., Dewell, S. & Papavasiliou, F. N. Transcriptome-wide sequencing reveals numerous APOBEC1 mRNA-editing targets in transcript 3’ UTRs. Nat. Struct. Mol. Biol. 18, 230–236 (2011).
Nishikura, K. A-to-I editing of coding and non-coding RNAs by ADARs. Nat. Rev. Mol. Cell Biol. 17, 83–96 (2016).
Bazak, L. et al. A-to-I RNA editing occurs at over a hundred million genomic sites, located in a majority of human genes. Genome Res. 24, 365–376 (2014).
Picardi, E. et al. Profiling RNA editing in human tissues: towards the inosinome Atlas. Sci. Rep. 5, 14941 (2015).
Mallela, A. & Nishikura, K. A-to-I editing of protein coding and noncoding RNAs. Crit. Rev. Biochem. Mol. Biol. 47, 493–501 (2012).
Mannion, N. M. et al. The RNA-editing enzyme ADAR1 controls innate immune responses to RNA. Cell Rep. 9, 1482–1494 (2014).
Gallo, A., Vukic, D., Michalik, D., O’Connell, M. A. & Keegan, L. P. ADAR RNA editing in human disease; more to it than meets the I. Hum. Genet. 136, 1265–1278 (2017).
Silvestris, D. A. et al. Dynamic inosinome profiles reveal novel patient stratification and gender-specific differences in glioblastoma. Genome Biol. 20, 33 (2019).
Ramaswami, G. et al. Accurate identification of human Alu and non-Alu RNA editing sites. Nat. Methods 9, 579–581 (2012).
Eisenberg, E. Bioinformatic approaches for identification of A-to-I editing sites. Curr. Top. Microbiol. Immunol. 353, 145–162 (2012).
Diroma, M. A., Ciaccia, L., Pesole, G. & Picardi, E. Elucidating the editome: bioinformatics approaches for RNA editing detection. Brief. Bioinform. 20, 436–447 (2019).
Nigita, G., Alaimo, S., Ferro, A., Giugno, R. & Pulvirenti, A. Knowledge in the investigation of A-to-I RNA editing signals. Front. Bioeng. Biotechnol. 3, 18 (2015).
Nigita, G. et al. ncRNA editing: functional characterization and computational resources. in Computational Biology of Non-Coding RNA (eds Lai, X., Gupta, S. K. & Vera, J.) 133–174 (Humana Press, 2019).
Picardi, E. & Pesole, G. REDItools: high-throughput RNA editing detection made easy. Bioinformatics 29, 1813–1814 (2013).
Picardi, E., D’Erchia, A. M., Lo Giudice, C. & Pesole, G. REDIportal: a comprehensive database of A-to-I RNA editing events in humans. Nucleic Acids Res. 45, D750–D757 (2017).
1000 Genomes Project Consortium. A global reference for human genetic variation. Nature 526, 68 (2015).
Chen, S., Zhou, Y., Chen, Y. & Gu, J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, 6 (2018).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Wang, M. & Kong, L. pblat: a multithread blat algorithm speeding up aligning sequences to genomes. BMC Bioinformatics 20, 28 (2019).
Kent, W. J. BLAT-the BLAST-like alignment tool. Genome Res. 12, 656–664 (2002).
Ross, C. A. & Tabrizi, S. J. Huntington’s disease: from molecular pathogenesis to clinical treatment. Lancet Neurol. 10, 15 (2011).
Hodges, A. et al. Regional and cellular gene expression changes in human Huntington’s disease brain. Hum. Mol. Genet. 15, 965–977 (2006).
Valor, L. M. Transcription, epigenetics and ameliorative strategies in Huntington’s Disease: a genome-wide perspective. Mol. Neurobiol. 51, 406–423 (2015).
Marti, E. et al. A myriad of miRNA variants in control and Huntington’s disease brain regions detected by massively parallel sequencing. Nucleic Acids Res. 38, 7219–7235 (2010).
Luthi-Carter, R. et al. Decreased expression of striatal signaling genes in a mouse model of Huntington’s disease. Hum. Mol. Genet. 9, 1259–1271 (2000).
Lin, L. et al. Transcriptome sequencing reveals aberrant alternative splicing in Huntington’s disease. Hum. Mol. Genet. 25, 3454–3466 (2016).
Annese, A. et al. Whole transcriptome profiling of late-onset Alzheimer’s disease patients provides insights into the molecular changes involved in the disease. Sci. Rep. 8, 4282 (2018).
D’Erchia, A. M. et al. Massive transcriptome sequencing of human spinal cord tissues provides new insights into motor neuron degeneration in ALS. Sci. Rep. 7, 10046 (2017).
Khermesh, K. et al. Reduced levels of protein recoding by A-to-I RNA editing in Alzheimer’s disease. RNA 22, 290–302 (2016).
Srivastava, P. K. et al. Genome-wide analysis of differential RNA editing in epilepsy. Genome Res. 27, 440–450 (2017).
Wang, L., Wang, S. & Li, W. RSeQC: quality control of RNA-seq experiments. Bioinformatics 28, 2184–2185 (2012).
DeLuca, D. S. et al. RNA-SeQC: RNA-seq metrics for quality control and process optimization. Bioinformatics 28, 1530–1532 (2012).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 10–12 (2011).
Patel, R. K. & Jain, M. NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PlOS One 7, e30619 (2012).
Wu, T. D. & Nacu, S. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics 26, 873–881 (2011).
Kim, D., Langmead, B. & Salzberg, S. L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360 (2015).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 2 (2012).
Yu, C. et al. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics 25, 2 (2009).
John, D., Weirick, T., Dimmeler, S. & Uchida, S. RNAEditor: easy detection of RNA editing events and the introduction of editing islands. Brief. Bioinform. 18, 8 (2012).
Wang, Z. et al. RES-Scanner: a software package for genome-wide identification of RNA-editing sites. Gigascience 5, 37 (2016).
Zhang, Q. & Xiao, X. Genome sequence-independent identification of RNA editing sites. Nat. Methods 12, 347 (2015).
Piechotta, M., Wyler, E., Ohler, U., Landthaler, M. & Dieterich, C. JACUSA: site-specific identification of RNA editing events from replicate sequencing data. BMC Bioinform. 18, 7 (2017).
Kim, M. S., Hur, B. & Kim, S. RDDpred: a condition-specific RNA-editing prediction model from RNA-seq data. BMC Genomics 17(Suppl 1)), 5 (2016).
Xiong, H. et al. RED-ML: a novel, effective RNA editing detection method based on machine learning. Gigascience https://doi.org/10.1093/gigascience/gix012 (2017).
Ouyang, Z. et al. Accurate identification of RNA editing sites from primitive sequence with deep neural networks. Sci. Rep. 8, 6005 (2018).
Porath, H. T., Carmi, S. & Levanon, E. Y. A genome-wide map of hyper-edited RNA reveals numerous new sites. Nat. Commun. 5, 4726 (2014).
Zhang, F., Lu, Y., Yan, S., Xing, Q. & Tian, W. SPRINT: an SNP-free toolkit for identifying RNA editing sites. Bioinformatics 33, 3538–3548 (2017).
Kiran, A. M., O’Mahony, J. J., Sanjeev, K. & Baranov, P. V. Darned in 2013: inclusion of model organisms and linking with Wikipedia. Nucleic Acids Res. 41, D258–261 (2013).
Ramaswami, G. & Li, J. B. RADAR: a rigorously annotated database of A-to-I RNA editing. Nucleic Acids Res. 42, D109–113 (2014).
Picardi, E., Horner, D. S. & Pesole, G. Single cell transcriptomics reveals specific RNA editing signatures in the human brain. RNA 23, 860–865 (2017).
Rossetti, C. et al. RNA editing signature during myeloid leukemia cell differentiation. Leukemia 31, 2824–2832 (2017).
Pinto, Y., Buchumenski, I., Levanon, E. Y. & Eisenberg, E. Human cancer tissues exhibit reduced A-to-I editing of miRNAs coupled with elevated editing of their targets. Nucleic Acids Res. 46, 71–82 (2018).
Lin, C.-H. & Chen, S. C.-C. The Cancer Editome Atlas: a resource for exploratory analysis of the adenosine-to-inosine RNA editome in cancer. Cancer Res. 79, 3001–3006 (2019).
Porath, H. T. et al. RNA editing is abundant and correlates with task performance in a social bumblebee. Nat. Commun. 10, 1605 (2019).
Liu, H. et al. A-to-I RNA editing is developmentally regulated and generally adaptive for sexual reproduction in Neurospora crassa. Proc. Natl Acad. Sci. USA 114, E7756–E7765 (2017).
Liew, Y. J., Li, Y., Baumgarten, S., Voolstra, C. R. & Aranda, M. Condition-specific RNA editing in the coral symbiont Symbiodinium microadriaticum. PLOS Genet. 13, e1006619 (2017).
Sapiro, A. L. et al. Illuminating spatial A-to-I RNA editing signatures within the Drosophila brain. Proc. Natl Acad. Sci. USA 116, 2318–2327 (2019).
Picardi, E. et al. Large-scale detection and analysis of RNA editing in grape mtDNA by RNA deep-sequencing. Nucleic Acids Res. 38, 4755–4767 (2010).
Wu, B. et al. Identification of symmetrical RNA editing events in the mitochondria of Salvia miltiorrhiza by strand-specific RNA sequencing. Sci. Rep. 7, 42250 (2017).
Picardi, E., D’Erchia, A. M., Montalvo, A. & Pesole, G. Using REDItools to detect RNA editing events in NGS datasets. Curr. Protoc. Bioinforma. 49, 12.12.1–12.12.15 (2015).
Picardi, E., D’Erchia, A. M., Gallo, A., Montalvo, A. & Pesole, G. Uncovering RNA editing sites in long non-coding RNAs. Front. Bioeng. Biotechnol. 2, 64 (2014).
Pinto, Y., Cohen, H. Y. & Levanon, E. Y. Mammalian conserved ADAR targets comprise only a small fragment of the human editosome. Genome Biol. 15, R5 (2014).
Deininger, P. Alu elements: know the SINEs. Genome Biol. 12, 236 (2011).
Porath, H. T., Knisbacher, B. A., Eisenberg, E. & Levanon, E. Y. Massive A-to-I RNA editing is common across the Metazoa and correlates with dsRNA abundance. Genome Biol. 18, 185 (2017).
Tran, S. S. et al. Widespread RNA editing dysregulation in brains from autistic individuals. Nat. Neurosci. 22, 25 (2019).
Wang, K., Li, M. & Hakonarson, H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38, e164 (2010).
Yang, H. & Wang, K. Genomic variant annotation and prioritization with ANNOVAR and wANNOVAR. Nat. Protoc. 10, 1556 (2015).
Tan, M. H. et al. Dynamic landscape and regulation of RNA editing in mammals. Nature 550, 249 (2017).
Acknowledgements
We kindly thank the ReCaS computing center at University of Bari for computational and technical assistance. We also acknowledge the PRACE project 2016163924 for computing resources. This work was supported by Elixir IIB and PRACE projects 2016163924 and 2018194670.
Author information
Authors and Affiliations
Contributions
G.P. and E.P. conceived the project. C.L.G. and E.P. designed and wrote the full analysis pipeline. C.L.G. performed extensive user testing of the software. M.A.T. created the docker image and implemented the statistical tests to calculate differential RNA editing. E.P. wrote the manuscript with input and editing from all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Protocols thanks Trees-Juen Chuang, Erez Levanon and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key references using this protocol
D’Erchia, A. M. et al. Sci. Rep. 7, 10046 (2017): https://doi.org/10.1038/s41598-017-10488-7
Picardi, E., Horner, D. S. and Pesole, G. RNA 23, 860–865 (2017): https://doi.org/10.1261/rna.058271.116
Rossetti, C. et al. Leukemia 31, 2824–2832 (2017): https://doi.org/10.1038/leu.2017.134
Supplementary information
Rights and permissions
About this article
Cite this article
Lo Giudice, C., Tangaro, M.A., Pesole, G. et al. Investigating RNA editing in deep transcriptome datasets with REDItools and REDIportal. Nat Protoc 15, 1098–1131 (2020). https://doi.org/10.1038/s41596-019-0279-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-019-0279-7
This article is cited by
-
RNA editing enzymes: structure, biological functions and applications
Cell & Bioscience (2024)
-
Transcriptomic analysis reveals associations of blood-based A-to-I editing with Parkinson’s disease
Journal of Neurology (2024)
-
FLARE: a fast and flexible workflow for identifying RNA editing foci
BMC Bioinformatics (2023)
-
Deep transcriptome profiling reveals limited conservation of A-to-I RNA editing in Xenopus
BMC Biology (2023)
-
L-GIREMI uncovers RNA editing sites in long-read RNA-seq
Genome Biology (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.