Abstract
Limited methods exist to assay the direct effects of therapeutic intervention on muscle stem cell fate, proliferation or differentiation in an in vivo context. Here we provide an optimized protocol for muscle stem cell isolation and transplantation into mice to deconvolute heterogeneity within isolated stem cell populations. Viable and pure cell populations are isolated within 2 h and can then be used for therapeutic intervention or transplantation to uncover the repopulating and differentiation potential in mice, a physiologically relevant in vivo context. Effects can be assessed 9 d after transplantation. This methodology analyzes cell and sort purity prior to transplantation to improve reproducibility and outlines novel blocking steps to improve tissue staining and analysis. Experience with surgical procedures in mice is recommended before attempting this protocol. Our system is widely applicable for exploring stem cell dynamics within muscle and has already been used to study heterogeneity within muscle stem cell populations and efficacy of therapeutic intervention on isolated stem cell populations.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Kuang, S., Kuroda, K., Le Grand, F. & Rudnicki, M. A. Asymmetric self-renewal and commitment of satellite stem cells in muscle. Cell 129, 999–1010 (2007).
Feige, P., Brun, C. E., Ritso, M. & Rudnicki, M. A. Orienting muscle stem cells for regeneration in homeostasis, aging and disease. Cell Stem Cell 23, 653–664 (2018).
Porpiglia, E. et al. High-resolution myogenic lineage mapping by single-cell mass cytometry. Nat. Cell Biol. 19, 558–567 (2017).
Giordani, L. et al. High-dimensional single-cell cartography reveals novel skeletal muscle-resident cell populations. Mol. Cell 74, 609–621.e6 (2019).
Sacco, A., Doyonnas, R., Kraft, P., Vitorovic, S. & Blau, H. M. Self-renewal and expansion of single transplanted muscle stem cells. Nature 456, 502–506 (2008).
Bar-Nur, O. et al. Direct reprogramming of mouse fibroblasts into functional skeletal muscle progenitors. Stem Cell Rep. 10, 1505–1521 (2018).
Zismanov, V. et al. Phosphorylation of eIF2α is a translational control mechanism regulating muscle stem cell quiescence and self-renewal. Cell Stem Cell 18, 79–90 (2016).
Wang, Y. X. et al. EGFR-Aurka signaling rescues polarity and regeneration defects in dystrophin-deficient muscle stem cells by increasing asymmetric divisions. Cell Stem Cell 24, 419–432.e6 (2019).
Pasut, A., Oleynik, P. & Rudnicki, M. A. Isolation of muscle stem cells by fluorescence activated cell sorting cytometry. Methods Mol. Biol. 798, 53–64 (2012).
Bentzinger, C. F., Wang, Y. X., Dumont, N. A. & Rudnicki, M. A. Cellular dynamics in the muscle satellite cell niche. EMBO Rep. 14, 1062–1072 (2013).
Gilbert, P. M. et al. Substrate elasticity regulates skeletal muscle stem cell self-renewal in culture. Science 329, 1078–1081 (2010).
Beauchamp, J. R., Morgan, J. E., Pagel, C. N. & Partridge, T. A. Dynamics of myoblast transplantation reveal a discrete minority of precursors with stem cell-like properties as the myogenic source. J. Cell Biol. 144, 1113–1122 (1999).
Qu, Z. et al. Development of approaches to improve cell survival in myoblast transfer therapy. J. Cell Biol. 142, 1257–1267 (1998).
Rocheteau, P., Gayraud-Morel, B., Siegl-Cachedenier, I., Blasco, M. A. & Tajbakhsh, S. A subpopulation of adult skeletal muscle stem cells retains all template DNA strands after cell division. Cell 148, 112–125 (2012).
Tierney, M. T., Stec, M. J., Rulands, S., Simons, B. D. & Sacco, A. Muscle stem cells exhibit distinct clonal dynamics in response to tissue repairand homeostatic aging. Cell Stem Cell 22, 119–127.e3 (2018).
Seale, P. et al. Pax7 is required for the specification of myogenic satellite cells. Cell 102, 777–786 (2000).
Von Maltzahn, J., Jones, A. E., Parks, R. J. & Rudnicki, M. A. Pax7 is critical for the normal function of satellite cells in adult skeletal muscle. Proc. Natl Acad. Sci. USA 110, 16474–16479 (2013).
Dumont, N. A. et al. Dystrophin expression in muscle stem cells regulates their polarity and asymmetric division. Nat. Med. 21, 1455–1463 (2015).
Machado, L. et al. In situ fixation redefines quiescence and early activation of skeletal muscle stem cells. Cell Rep. 21, 1982–1993 (2017).
Van Velthoven, C. T. J., de Morree, A., Egner, I. M., Brett, J. O. & Rando, T. A. Transcriptional profiling of quiescent muscle stem cells in vivo. Cell Rep. 21, 1994–2004 (2017).
Van den Brink, S. C. et al. Single-cell sequencing reveals dissociation-induced gene expression in tissue subpopulations. Nat. Methods 14, 935–936 (2017).
Lepper, C., Partridge, T. A. & Fan, C. M. An absolute requirement for Pax7-positive satellite cells in acute injury-induced skeletal muscle regeneration. Development 138, 3639–3646 (2011).
Price, F. D. et al. Inhibition of JAK/STAT signaling stimulates adult satellite cell function. Nat. Med. 20, 1174–1181 (2014).
Judson, R. N. et al. Inhibition of methyltransferase Setd7 allows the in vitro expansion of myogenic stem cells with improved therapeutic potential. Cell Stem Cell 22, 177–190 (2018).
Moreno-Garcia, A., Kun, A., Calero, O., Medina, M. & Calero, M. An overview of the role of lipofuscin in age-related neurodegeneration. Front. Neurosci. 12, 464 (2018).
Acknowledgements
P.F. is supported by a fellowship from Canadian Institutes for Health Research. M.A.R. holds the Canada Research Chair in Molecular Genetics. These studies were carried out with grant support to M.A.R. from the NIH (R01AR044031), the Canadian Institutes for Health Research (FDN-148387), E-Rare-2: Canadian Institutes of Health Research/Muscular Dystrophy Canada (ERA-132935), the Muscular Dystrophy Association, the Ontario Institute for Regenerative Medicine and the Stem Cell Network.
Author information
Authors and Affiliations
Contributions
Conceptualization, P.F. and M.A.R.; methodology, P.F. and M.A.R.; writing, P.F. and M.A.R.; funding acquisition, M.A.R.; resources, M.A.R; supervision, M.A.R.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Protocols thanks Robert W. Arpke, Dawn Cornelison and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key references that demonstrate the development/use of the Protocol
Wang, Y. X. et al. Cell Stem Cell. 24, 419–432.e6 (2019): https://doi.org/10.1016/j.stem.2019.01.002
Bentzinger, C. F. et al. Cell Stem Cell. 12, 75–87 (2013): https://doi.org/10.1016/j.stem.2012.09.015
Kuang, S., Kuroda, K., Le Grand, F., & Rudnicki, M. A. Cell. 129, 999–1010 (2007): https://doi.org/10.1016/j.cell.2007.03.044
Integrated supplementary information
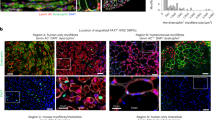
Supplementary Fig. 1 Cell purity quality control prior to engraftment.
Representative immunofluorescence images with single channels for committed satellite cell transplants (a) and satellite stem cell transplants (b). Blue, DAPI; green, nGFP; red, nTdTomato; white, Pax7. Scale bar, 50 μm. n = 3, mean ± s.d. Experimental protocols for mice used in this study were performed in accordance with the guidelines established by the University of Ottawa Animal Care Committee, which is based on the guidelines of the Canadian Council on Animal Care.
Supplementary Fig. 2 Lipofuscin-quenching validation for transplanted tissue.
a, Representative immunofluorescence images of non-injured, non-transplanted muscle show the high levels of autofluorescence present within skeletal muscle. A trace of the 12 Bit histogram from Zen 2.5 for 488, 546 and 647 nm is shown. b, Lipofuscin-quenched sections from serial sections have significantly decreased background in 488 and 546 nm autofluorescence with a slight increase in 647 nm autofluorescence. c and d, Representative immunofluorescence images from satellite stem cell (nTdTomato) transplants treated without TrueBlack (c) show the significant presence of non-cellular (DAPI-negative) autofluorescence artifact within injured skeletal muscle, which impedes the ability to detect real signal from transplanted cells as shown following blocking with TrueBlack (d). Blue, DAPI; green, nGFP; red, nTdTomato; white, Pax7. Scale bar, 100 μm. Experimental protocols for mice used in this study were performed in accordance with the guidelines established by the University of Ottawa Animal Care Committee, which is based on the guidelines of the Canadian Council on Animal Care.
Supplementary information
Supplementary Information
Supplementary Figs. 1 and 2.
Rights and permissions
About this article
Cite this article
Feige, P., Rudnicki, M.A. Isolation of satellite cells and transplantation into mice for lineage tracing in muscle. Nat Protoc 15, 1082–1097 (2020). https://doi.org/10.1038/s41596-019-0278-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-019-0278-8
This article is cited by
-
Analysis of human satellite cell dynamics on cultured adult skeletal muscle myofibers
Skeletal Muscle (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.