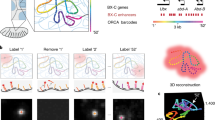
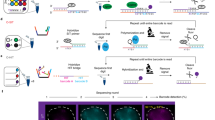
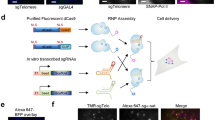
Abstract
Simultaneous observation of 3D chromatin organization and transcription at the single-cell level and with high spatial resolution may hold the key to unveiling the mechanisms regulating embryonic development, cell differentiation and even disease. We recently developed Hi-M, a technology that enables the sequential labeling, 3D imaging and localization of multiple genomic DNA loci, together with RNA expression, in single cells within whole, intact Drosophila embryos. Importantly, Hi-M enables simultaneous detection of RNA expression and chromosome organization without requiring sample unmounting and primary probe rehybridization. Here, we provide a step-by-step protocol describing the design of probes, the preparation of samples, the stable immobilization of embryos in microfluidic chambers, and the complete procedure for image acquisition. The combined RNA/DNA fluorescence in situ hybridization procedure takes 4–5 d, including embryo collection. In addition, we describe image analysis software to segment nuclei, detect genomic spots, correct for drift and produce Hi-M matrices. A typical Hi-M experiment takes 1–2 d to complete all rounds of labeling and imaging and 4 additional days for image analysis. This technology can be easily expanded to investigate cell differentiation in cultured cells or organization of chromatin within complex tissues.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout











Similar content being viewed by others
Data and code availability
The code used and described in this paper and the experimental dataset used to construct Fig. 11 have been uploaded to https://doi.org/10.17632/5f5hd9yj3z.1#folder-26d1f8c0-fc58-4b87-8c4f-cd8a294a555Software. Additional advice on how to use these can be obtained from the authors upon reasonable request. Further information and requests for resources, reagents and software should be directed to and will be fulfilled by the lead contact, M.N. (marcelo.nollmann@cbs.cnrs.fr).
References
Bonev, B. & Cavalli, G. Organization and function of the 3D genome. Nat. Rev. Genet. 17, 661–678 (2016). erratum 17, 772 (2016).
Giorgetti, L. & Heard, E. Closing the loop: 3C versus DNA FISH. Genome Biol. 17, 215 (2016).
Cardozo Gizzi, A. M. et al. Microscopy-based chromosome conformation capture enables simultaneous visualization of genome organization and transcription in intact organisms. Mol. Cell 74, 212–222.e5 (2019).
Bintu, B. et al. Super-resolution chromatin tracing reveals domains and cooperative interactions in single cells. Science 362, aau1783 (2018).
Nir, G. et al. Walking along chromosomes with super-resolution imaging, contact maps, and integrative modeling. PLoS Genet. 14, e1007872 (2018).
Mateo, L. J. et al. Visualizing DNA folding and RNA in embryos at single-cell resolution. Nature 568, 49–54 (2019).
Beliveau, B. J. et al. Versatile design and synthesis platform for visualizing genomes with Oligopaint FISH probes. Proc. Natl Acad. Sci. USA 109, 21301–21306 (2012).
Beliveau, B. J. et al. Single-molecule super-resolution imaging of chromosomes and in situ haplotype visualization using Oligopaint FISH probes. Nat. Commun. 6, 7147 (2015).
Lubeck, E., Coskun, A. F., Zhiyentayev, T., Ahmad, M. & Cai, L. Single-cell in situ RNA profiling by sequential hybridization. Nat. Methods 11, 360–361 (2014).
Chen, K. H., Boettiger, A. N., Moffitt, J. R., Wang, S. & Zhuang, X. RNA imaging. Spatially resolved, highly multiplexed RNA profiling in single cells. Science 348, aaa6090 (2015).
Moffitt, J. R. et al. Molecular, spatial, and functional single-cell profiling of the hypothalamic preoptic region. Science 362, aau5324 (2018).
Moffitt, J. R. et al. High-performance multiplexed fluorescence in situ hybridization in culture and tissue with matrix imprinting and clearing. Proc. Natl Acad. Sci. USA 113, 14456–14461 (2016).
Rosin, L. F., Nguyen, S. C. & Joyce, E. F. Condensin II drives large-scale folding and spatial partitioning of interphase chromosomes in Drosophila nuclei. PLoS Genet. 14, e1007393 (2018).
Kishi, J. Y., Beliveau, B. J., Lapan, S. W., West, E. R. & Zhu, A. SABER enables highly multiplexed and amplified detection of DNA and RNA in cells and tissues. bioRxiv (2018).
Fields, B. D., Nguyen, S. C., Nir, G. & Kennedy, S. A multiplexed DNA FISH strategy for assessing genome architecture in Caenorhabditis elegans. Elife 8, e42823 (2019).
Pallikkuth, S. et al. Sequential super-resolution imaging using DNA strand displacement. PLoS One 13, e0203291 (2018).
Roohi, J., Cammer, M., Montagna, C. & Hatchwell, E. An improved method for generating BAC DNA suitable for FISH. Cytogenet. Genome Res. 121, 7–9 (2008).
Bienko, M. et al. A versatile genome-scale PCR-based pipeline for high-definition DNA FISH. Nat. Methods 10, 122–124 (2013).
Wang, S. et al. Spatial organization of chromatin domains and compartments in single chromosomes. Science 353, 598–602 (2016).
Shah, S. et al. Dynamics and spatial genomics of the nascent transcriptome by intron seqFISH. Cell 174, 363–376.e16 (2018).
Eng, C.-H. L., Shah, S., Thomassie, J. & Cai, L. Profiling the transcriptome with RNA SPOTs. Nat. Methods 14, 1153–1155 (2017).
Wu, X., Mao, S., Ying, Y., Krueger, C. J. & Chen, A. K. Progress and challenges for live-cell imaging of genomic loci using CRISPR-based platforms. Genomics Proteomics Bioinformatics 17, 119–128 (2019).
Ma, H. et al. Multiplexed labeling of genomic loci with dCas9 and engineered sgRNAs using CRISPRainbow. Nat. Biotechnol. 34, 528–530 (2016).
Germier, T. et al. Real-time imaging of a single gene teveals transcription-initiated local confinement. Biophys. J. 113, 1383–1394 (2017).
Chen, H. et al. Dynamic interplay between enhancer–promoter topology and gene activity. Nat. Genet. 50, 1296–1303 (2018).
Yunger, S., Rosenfeld, L., Garini, Y. & Shav-Tal, Y. Single-allele analysis of transcription kinetics in living mammalian cells. Nat. Methods 7, 631–653 (2010).
Larson, D. R., Zenklusen, D., Wu, B., Chao, J. A. & Singer, R. H. Real-time observation of transcription initiation and elongation on an endogenous yeast gene. Science 332, 475–478 (2011).
Fukaya, T., Lim, B. & Levine, M. Enhancer control of transcriptional bursting. Cell 166, 358–368 (2016).
Saad, H. et al. DNA dynamics during early double-strand break processing revealed by non-intrusive imaging of living cells. PLoS Genet. 10, e1004187 (2014).
Boyle, S., Rodesch, M. J., Halvensleben, H. A., Jeddeloh, J. A. & Bickmore, W. A. Fluorescence in situ hybridization with high-complexity repeat-free oligonucleotide probes generated by massively parallel synthesis. Chromosome Res. 19, 901–909 (2011).
Rouillard, J.-M., Zuker, M. & Gulari, E. OligoArray 2.0: design of oligonucleotide probes for DNA microarrays using a thermodynamic approach. Nucleic Acids Res 31, 3057–3062 (2003).
Gelali, E. et al. iFISH is a publically available resource enabling versatile DNA FISH to study genome architecture. Nat. Commun. 10, 1636 (2019).
Beliveau, B. J. et al. OligoMiner provides a rapid, flexible environment for the design of genome-scale oligonucleotide in situ hybridization probes. Proc. Natl Acad. Sci. USA 115, E2183–E2192 (2018).
Raj, A., van den Bogaard, P., Rifkin, S. A., van Oudenaarden, A. & Tyagi, S. Imaging individual mRNA molecules using multiple singly labeled probes. Nat. Methods 5, 877–879 (2008).
Moffitt, J. R. & Zhuang, X. RNA imaging with multiplexed error-robust fluorescence in situ hybridization (MERFISH). Methods Enzymol. 572, 1–49 (2016).
Trcek, T., Lionnet, T., Shroff, H. & Lehmann, R. mRNA quantification using single-molecule FISH in Drosophila embryos. Nat. Protoc. 12, 1326–1348 (2017).
Shpiz, S., Lavrov, S. & Kalmykova, A. Combined RNA/DNA fluorescence in situ hybridization on whole-mount Drosophila ovaries. Methods Mol. Biol. 1093, 161–169 (2014).
Boettiger, A. N. & Levine, M. Rapid transcription fosters coordinate snail expression in the Drosophila embryo. Cell Rep. 3, 8–15 (2013).
Bantignies, F. & Cavalli, G. Topological organization of Drosophila Hox genes using DNA fluorescent in situ hybridization. Methods Mol. Biol. 1196, 103–120 (2014).
Moffitt, J. R. et al. High-throughput single-cell gene-expression profiling with multiplexed error-robust fluorescence in situ hybridization. Proc. Natl Acad. Sci. USA 113, 11046–11051 (2016).
Fung, J. C., Marshall, W. F., Dernburg, A., Agard, D. A. & Sedat, J. W. Homologous chromosome pairing in Drosophila melanogaster proceeds through multiple independent initiations. J. Cell Biol. 141, 5–20 (1998).
Cattoni, D. I. et al. Single-cell absolute contact probability detection reveals chromosomes are organized by multiple low-frequency yet specific interactions. Nat. Commun. 8, 1753 (2017).
Lagha, M. et al. Paused Pol II coordinates tissue morphogenesis in the Drosophila embryo. Cell 153, 976–987 (2013).
Ashburner, M., Golic, K. G. & Hawley, R. S. Drosophila: A Laboratory Handbook (Cold Spring Harbor Laboratory Press, 2011).
Ferrandiz, C. & Sessions, A. Preparation and hydrolysis of digoxygenin-labeled probes for in situ hybridization of plant tissues. CSH Protoc. https://doi.org/10.1101/pdb.prot4942 (2008).
Acknowledgements
This project received funding from the European Research Council (ERC) under the European Union’s Horizon 2020 Research and Innovation Program (grant 724429). This work also benefited from support from Labex EpiGenMed, an ‘Investments for the Future’ program (grant ANR-10-LABX-12-01). We acknowledge the France-BioImaging infrastructure supported by the French National Research Agency (grant ANR-10-INBS-04, ‘Investments for the Future’).
Author information
Authors and Affiliations
Contributions
A.M.C.G. and M.N. designed the experiments. A.M.C.G. and M.N. designed the Oligopaints probes. C.H. amplified and purified the Oligopaints libraries. A.M.C.G., C.H., S.M.E. and J.G. developed the RNA/DNA staining protocol; A.M.C.G., S.M.E., J.G. and D.I.C. conducted the experiments. J.-B.F. designed and built the microscopy setup and acquisition software. M.N. developed the software for image analysis. A.M.C.G., and J.G. analyzed the data. A.M.C.G., D.I.C. and M.N. wrote the manuscript. All authors reviewed and commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related link
Key reference using this protocol
Cardozo Gizzi, A. M. et al. Mol. Cell 74, 212–222.e5 (2019): https://doi.org/10.1016/j.molcel.2019.01.011
Integrated supplementary information
Supplementary Figure 1 Microfluidic setup plan.
Schematic representation of microfluidics used in Hi-M setup. Buffers and readout probes are selected using a combination of three eight-way HVXM8-5 valves connected in series. The fluid handling circuit consist of 21 tubes connected each to one of the valves. Hi-M wash buffer and 2× SSC pass through an online degassing unit before connecting to the corresponding valve. The other 19 valves are connected to 17 readout solution tubes, the chemical bleaching buffer and the acquisition solution. The sample is mounted to the FCS2 flow chamber. Continuous flow is created by a negative-pressure pump. Before starting the experiment, the system is fill with 2× SCC buffer, assuring the removal of all air bubbles. Microfluidics is automatically controlled using a homemade LabView 2015 software. Image adapted with permission from ref. 3, Elsevier.
Supplementary information
Supplementary Information
Supplementary Fig. 1 and Supplementary Tables 2–4.
Supplementary Table 1
Oligo pool sequences
Rights and permissions
About this article
Cite this article
Cardozo Gizzi, A.M., Espinola, S.M., Gurgo, J. et al. Direct and simultaneous observation of transcription and chromosome architecture in single cells with Hi-M. Nat Protoc 15, 840–876 (2020). https://doi.org/10.1038/s41596-019-0269-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-019-0269-9
This article is cited by
-
pyHiM: a new open-source, multi-platform software package for spatial genomics based on multiplexed DNA-FISH imaging
Genome Biology (2024)
-
Polycomb repression of Hox genes involves spatial feedback but not domain compaction or phase transition
Nature Genetics (2024)
-
The spatial organization of transcriptional control
Nature Reviews Genetics (2023)
-
3D chromatin interactions involving Drosophila insulators are infrequent but preferential and arise before TADs and transcription
Nature Communications (2023)
-
Simultaneous profiling of chromatin architecture and transcription in single cells
Nature Structural & Molecular Biology (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.