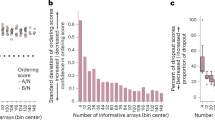
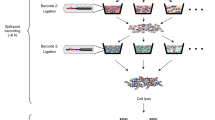
Abstract
Temporally resolved assays of bacterial gene expression using printed fluorescence imaging boxes (PFIboxes) are non-destructive, inexpensive and simple to prepare. Herein, we describe a full experimental pipeline wherein PFIbox parts are modified and 3D printed, electronics assembled and used to study transcriptional responses of Escherichia coli to chemical stressors. A chemical probe is added to agar growth medium, and a promoter–fluorophore fusion library is arrayed in high density on the agar slab. With high temporal resolution, the reporter library is imaged in PFIboxes, then quantified using promoter activity as a measure of gene expression. PFIboxes have advantages over conventional transcriptomic approaches such as RNA-seq, as the non-destructive nature permits a high-resolution temporal dimension in the data. This results in rapid measurement of transcriptional responses to chemical or physical stimuli. Each time-course gene expression assay costs about US$2 to run, in triplicate, using this method. Printing time depends on printer and settings, but once printed, PFIboxes can be fully assembled, programmed and loaded with samples in less than 1 h. Experimental durations and sampling frequency are set according to user need, but can be run in the duration of a microbial growth curve.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout










Similar content being viewed by others
Data availability
All files listed in this protocol, including 3D models and image analysis macros, can be found on the PFIbox GitHub repository at https://github.com/sfrench007/pfibox. The raw data from Figs. 3, 9 and 10 can be found on Mendeley Data at https://doi.org/10.17632/6hjcdr3td3.1. The dataset for the French et al.4 PFIbox manuscript can also be found on Mendeley Data at https://doi.org/10.17632/fmv6vv4bsf.1. All files and data are freely available, and licensed under the BSD-3 clause.
Code availability
Analysis code can be found on the PFIbox GitHub repository at https://github.com/sfrench007/pfibox. The code in this protocol has been peer reviewed.
References
Massé, E., Escorcia, F. E. & Gottesman, S. Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes Dev. 17, 2374–2383 (2003).
Hausser, J., Mayo, A., Keren, L. & Alon, U. Central dogma rates and the trade-off between precision and economy in gene expression. Nat. Commun. 10, 68 (2019).
Mok, W. W. K. & Brynildsen, M. P. Timing of DNA damage responses impacts persistence to fluoroquinolones. Proc. Natl Acad. Sci. USA 115, E6301–E6309 (2018).
French, S., Coutts, B. E. & Brown, E. D. Open-source high-throughput phenomics of bacterial promoter-reporter strains. Cell Syst. 7, 339–346.e3 (2018).
Nuñez, I. et al. Low cost and open source multi-fluorescence imaging system for teaching and research in biology and bioengineering. PLoS ONE 12, e0187163 (2017).
MacNair, C. R. et al. A cell-based approach to characterize antimicrobial compounds through kinetic dose response. Bioorg. Med. Chem. 24, 6315–6319 (2016).
French, S. et al. A robust platform for chemical genomics in bacterial systems. Mol. Biol. Cell 27, 1015–1025 (2016).
Nichols, R. J. et al. Phenotypic landscape of a bacterial cell. Cell 144, 143–156 (2011).
Tamae, C. et al. Determination of antibiotic hypersensitivity among 4,000 single-gene-knockout mutants of Escherichia coli. J. Bacteriol. 190, 5981–5988 (2008).
Creecy, J. P. & Conway, T. Quantitative bacterial transcriptomics with RNA-seq. Curr. Opin. Microbiol. 23, 133–140 (2015).
Hör, J., Gorski, S. A. & Vogel, J. Bacterial RNA biology on a genome scale. Mol. Cell 70, 785–799 (2018).
Leppek, K. & Barna, M. An rRNA variant to deal with stress. Nat. Microbiol 4, 382–383 (2019).
Song, W. et al. Divergent rRNAs as regulators of gene expression at the ribosome level. Nat. Microbiol. 4, 515–526 (2019).
Aprianto, R., Slager, J., Holsappel, S. & Veening, J.-W. High-resolution analysis of the pneumococcal transcriptome under a wide range of infection-relevant conditions. Nucleic Acids Res 46, 9990–10006 (2018).
Zaslaver, A. et al. A comprehensive library of fluorescent transcriptional reporters for Escherichia coli. Nat. Methods 3, 623–628 (2006).
Keseler, I. M. et al. EcoCyc: a comprehensive database of Escherichia coli biology. Nucleic Acids Res. 39, D583–D590 (2011).
Keseler, I. M. et al. EcoCyc: fusing model organism databases with systems biology. Nucleic Acids Res. 41, D605–D612 (2012).
Jaeger, P. A., McElfresh, C., Wong, L. R. & Ideker, T. Beyond agar: gel substrates with improved optical clarity and drug efficiency and reduced autofluorescence for microbial growth experiments. Appl. Environ. Microbiol. 81, 5639–5649 (2015).
Schneider, C. A., Rasband, W. S. & Eliceiri, K. W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Bongaerts, R. J. M., Hautefort, I., Sidebotham, J. M. & Hinton, J. C. D. Green fluorescent protein as a marker for conditional gene expression in bacterial cells. Methods Enzymol. 358, 43–66 (2002).
Kumar, L. & Futschik, M. E. Mfuzz: a software package for soft clustering of microarray data. Bioinformation 2, 5–7 (2012).
Karp, P. D. et al. Pathway tools version 19.0 update: software for pathway/genome informatics and systems biology. Brief. Bioinform. 17, 877–890 (2016).
Ashburner, M. et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25, 25–29 (2000).
Supek, F., Bošnjak, M., Škunca, N. & Šmuc, T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS ONE 6, e21800 (2011).
Ihaka, R. & Gentleman, R. R: a language for data analysis and graphics. J. Comput. Graph. Stat. 5, 299–314 (1996).
Mangat, C. S., Bharat, A., Gehrke, S. S. & Brown, E. D. Rank ordering plate data facilitates data visualization and normalization in high-throughput screening. J. Biomol. Screen. 19, 1314–1320 (2014).
Huber, W. et al. Orchestrating high-throughput genomic analysis with bioconductor. Nat. Methods 12, 115–21 (2015).
Futschik, M. E. & Carlisle, B. Noise-robust soft clustering of gene expression time-course data. J. Bioinform. Comput. Biol. 3, 965–988 (2005).
Acknowledgements
This research was supported by a Foundation grant from the Canadian Institutes for Health Research (FRN 143215), by the Natural Sciences and Engineering Research Council of Canada, by infrastructure funding from Canada Foundation for Innovation, and by a Tier I Canada Research Chair award to E.D.B. A.B.Y.G. was funded by an award from the Canadian Institutes for Health Research.
Author information
Authors and Affiliations
Contributions
S.F. designed and created the PFIbox hardware and software, and developed the protocol. A.B.Y.G. refined the software in the protocol, optimized hardware settings and ran the experimental pipeline to generate data. E.D.B. supervised the work, and provided guidance and direction on all components of the protocol.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Protocols thanks Geoff Baldwin, Jan-Willem Veening and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related link
Key reference using this protocol
French, S., Coutts, B. E. & Brown, E. D. Cell Systems 7, 339–346.e3 (2018): https://doi.org/10.1016/j.cels.2018.07.004
Supplementary information
Rights and permissions
About this article
Cite this article
French, S., Guo, A.B.Y. & Brown, E.D. A comprehensive guide to dynamic analysis of microbial gene expression using the 3D-printed PFIbox and a fluorescent reporter library. Nat Protoc 15, 575–603 (2020). https://doi.org/10.1038/s41596-019-0257-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-019-0257-0
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.