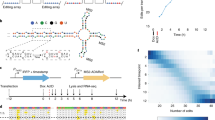
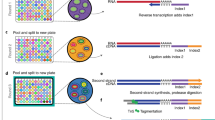
Abstract
It is difficult to elucidate the transcriptional history of a cell using current experimental approaches, as they are destructive in nature and therefore describe only a moment in time. To overcome these limitations, we recently established Record-seq, a technology that enables transcriptional recording by CRISPR spacer acquisition from RNA. The recorded transcriptomes are recovered by SENECA, a method that selectively amplifies expanded CRISPR arrays, followed by deep sequencing. The resulting CRISPR spacers are aligned to the host genome, thereby enabling transcript quantification and associated analyses. Here, we describe the experimental procedures of the Record-seq workflow as well as subsequent data analysis. Beginning with the experimental design, Record-seq data can be obtained and analyzed within 1–2 weeks.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
Deep sequencing data are available in the National Center for Biotechnology Information Sequence Read Archive (PRJNA510019). An example counts dataset can be accessed from https://github.com/plattlab/Transcriptional-Recording/tree/master/example-data.
Code availability
The python code is available in the Supplementary Data. The latest versions of the code can be accessed at the Platt lab github (https://github.com/plattlab/Transcriptional-Recording). The code in this protocol has been peer reviewed.
References
Schmidt, F. & Platt, R. J. Applications of CRISPR-Cas for synthetic biology and genetic recording. Curr. Opin. Syst. Biol. 5, 9–15 (2017).
Farzadfard, F. & Lu, T. K. Emerging applications for DNA writers and molecular recorders. Science 361, 870–875 (2018).
Esvelt, K. M. & Wang, H. H. Genome-scale engineering for systems and synthetic biology. Mol. Syst. Biol. 9, 641 (2013).
Farzadfard, F. & Lu, T. K. Synthetic biology. Genomically encoded analog memory with precise in vivo DNA writing in living cell populations. Science 346, 1256272 (2014).
Roquet, N., Soleimany, A. P., Ferris, A. C., Aaronson, S. & Lu, T. K. Synthetic recombinase-based state machines in living cells. Science 353, aad8559 (2016).
Weinberg, B. H. et al. Large-scale design of robust genetic circuits with multiple inputs and outputs for mammalian cells. Nat. Biotechnol. 35, 453–462 (2017).
Zamft, B. M. et al. Measuring cation dependent DNA polymerase fidelity landscapes by deep sequencing. PloS ONE 7, e43876 (2012).
McKenna, A. et al. Whole-organism lineage tracing by combinatorial and cumulative genome editing. Science 353, aaf7907 (2016).
Raj, B. et al. Simultaneous single-cell profiling of lineages and cell types in the vertebrate brain. Nat. Biotechnol. 36, 442–450 (2018).
Frieda, K. L. et al. Synthetic recording and in situ readout of lineage information in single cells. Nature 541, 107–111 (2017).
Perli, S. D., Cui, C. H. & Lu, T. K. Continuous genetic recording with self-targeting CRISPR-Cas in human cells. Science 353, https://doi.org/10.1126/science.aag0511 (2016).
Tang, W. & Liu, D. R. Rewritable multi-event analog recording in bacterial and mammalian cells. Science 360, https://doi.org/10.1126/science.aap8992 (2018).
Sheth, R. U., Yim, S. S., Wu, F. L. & Wang, H. H. Multiplex recording of cellular events over time on CRISPR biological tape. Science 358, 1457–1461 (2017).
Silas, S. et al. Direct CRISPR spacer acquisition from RNA by a natural reverse transcriptase-Cas1 fusion protein. Science 351, aad4234 (2016).
Shipman, S. L., Nivala, J., Macklis, J. D. & Church, G. M. Molecular recordings by directed CRISPR spacer acquisition. Science 353, aaf1175 (2016).
Shipman, S. L., Nivala, J., Macklis, J. D. & Church, G. M. CRISPR-Cas encoding of a digital movie into the genomes of a population of living bacteria. Nature 547, 345–349 (2017).
Schmidt, F., Cherepkova, M. Y. & Platt, R. J. Transcriptional recording by CRISPR spacer acquisition from RNA. Nature 562, 380–385 (2018).
Nuñez, J. K. et al. Cas1–Cas2 complex formation mediates spacer acquisition during CRISPR–Cas adaptive immunity. Nat. Struct. Mol. Biol. 21, 528–534 (2014).
Jackson, S. A. et al. CRISPR-Cas: adapting to change. Science 356, https://doi.org/10.1126/science.aal5056 (2017).
Yosef, I., Goren, M. G. & Qimron, U. Proteins and DNA elements essential for the CRISPR adaptation process in Escherichia coli. Nucleic Acids Res. 40, 5569–5576 (2012).
Koster, J. & Rahmann, S. Snakemake—a scalable bioinformatics workflow engine. Bioinformatics 28, 2520–2522 (2012).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Li, H. et al. The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Liao, Y., Smyth, G. K. & Shi, W. The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 41, e108 (2013).
Leinonen, R., Sugawara, H. & Shumway, M., International Nucleotide Sequence Database, C. The sequence read archive. Nucleic Acids Res. 39, D19–21 (2011).
Stead, M. B. et al. RNAsnap: a rapid, quantitative and inexpensive, method for isolating total RNA from bacteria. Nucleic Acids Res. 40, e156 (2012).
Acknowledgements
We thank C. Beisel, E. Burcklen, K. Eschbach, T. Schär, and I. Nissen from the Genomics Facility Basel for assistance in Illumina sequencing. F.S, M.Y.C, T.T., and R.J.P. are supported in part by funds from the Swiss National Science Foundation, ETH domain Personalized Health and Related Technologies, Brain and Behavior Research Foundation, and the National Centres of Competence–Molecular Systems Engineering. Plasmid reagents are available through Addgene. Correspondence and requests for materials should be addressed to R.J.P. (rplatt@ethz.ch).
Author information
Authors and Affiliations
Contributions
F.S., M.Y.C., T.T., and R.J.P. designed the study. F.S. performed experiments, T.T. and F.S. analyzed data, T.T. created the python pipeline for primary analysis, T.T. and M.O created the recoRdseq library for secondary analysis, F.S., T.T, M.Y.C., and R.J.P. wrote the manuscript with input from all authors.
Corresponding author
Ethics declarations
Competing interests
R.J.P. and F.S. are inventors on a patent application filed by ETH Zurich relating to work in this article. All other authors have no competing interests.
Additional information
Peer review information Nature Protocols thanks Stan J. J. Brouns, Simon Jackson and Malcolm F. White for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related link
Key references using this protocol
Schmidt, F. et al. Nature 562, 380–385 (2018): https://doi.org/10.1038/s41586-018-0569-1
Integrated supplementary information
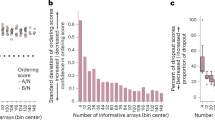
Supplementary Figure 1 Additional illustrative output plots generated by the recoRdseq R package, with samples color-coded by experimental group specified in the design matrix.
a. Mean untransformed genome-aligning spacer counts by group (error bars represent standard error of the mean). b. PCA plots for rlog transformed gene-aligning spacer counts for the top 50 variable genes with clusters defined by k-means clustering (k=2). c. Venn diagram showing overlap in significant DE genes (padj < 0.05) detected by the three DE tools (DESeq2, edgeR and baySeq). d. Heatmap showing unsupervised hierarchical clustering of samples based on rlog transformed genome-aligning spacer counts for high-confidence DE genes detected by the three DE tools. For all panels, n=6 independent biological replicates.
Supplementary information
Rights and permissions
About this article
Cite this article
Tanna, T., Schmidt, F., Cherepkova, M.Y. et al. Recording transcriptional histories using Record-seq. Nat Protoc 15, 513–539 (2020). https://doi.org/10.1038/s41596-019-0253-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-019-0253-4
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.