Abstract
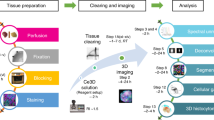
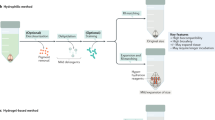
Tissue-clearing techniques are powerful tools for biological research and pathological diagnosis. Here, we describe advanced clear, unobstructed brain imaging cocktails and computational analysis (CUBIC) procedures that can be applied to biomedical research. This protocol enables preparation of high-transparency organs that retain fluorescent protein signals within 7–21 d by immersion in CUBIC reagents. A transparent mouse organ can then be imaged by a high-speed imaging system (>0.5 TB/h/color). In addition, to improve the understanding and simplify handling of the data, the positions of all detected cells in an organ (3–12 GB) can be extracted from a large image dataset (2.5–14 TB) within 3–12 h. As an example of how the protocol can be used, we counted the number of cells in an adult whole mouse brain and other distinct anatomical regions and determined the number of cells transduced with mCherry following whole-brain infection with adeno-associated virus (AAV)-PHP.eB. The improved throughput offered by this protocol allows analysis of numerous samples (e.g., >100 mouse brains per study), providing a platform for next-generation biomedical research.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
CUBIC-Atlas v.1.2 reference data are available from http://cubic-atlas.riken.jp. Other datasets generated or analyzed during the current study are available from the corresponding author upon reasonable request.
Code availability
All source code and a brief guide for data analysis are available on https://github.com/lsb-riken/CUBIC-informatics. Source code for the microscope and MOVIE system is available from the corresponding author upon reasonable request.
References
Voie, A. H., Burns, D. H. & Spelman, F. A. Orthogonal-plane fluorescence optical sectioning: three-dimensional imaging of macroscopic biological specimens. J. Micros. 170, 229–236 (1993).
Keller, P. J., Schmidt, A. D., Wittbrodt, J. & Stelzer, E. H. Reconstruction of zebrafish early embryonic development by scanned light sheet microscopy. Science 322, 1065–1069 (2008).
Huisken, J., Swoger, J., Del Bene, F., Wittbrodt, J. & Stelzer, E. H. K. Optical sectioning deep inside live embryos by selective plane illumination microscopy. Science 305, 1007–1009 (2004).
Dodt, H. U. et al. Ultramicroscopy: three-dimensional visualization of neuronal networks in the whole mouse brain. Nat. Methods 4, 331–336 (2007).
Kim, Y. et al. Mapping social behavior-induced brain activation at cellular resolution in the mouse. Cell Rep. 10, 292–305 (2015).
Tomer, R. et al. SPED light sheet microscopy: fast mapping of biological system structure and function. Cell 163, 1796–1806 (2015).
Kubota, S. I. et al. Whole-body profiling of cancer metastasis with single-cell resolution. Cell Rep. 20, 236–250 (2017).
Nojima, S. et al. CUBIC pathology: three-dimensional imaging for pathological diagnosis. Sci. Rep. 7, 9269 (2017).
Glaser, A. K. et al. Light-sheet microscopy for slide-free non-destructive pathology of large clinical specimens. Nat. Biomed. Eng. 1, 0084 (2017).
Tainaka, K. et al. Chemical landscape for tissue clearing based on hydrophilic reagents. Cell Rep. 24, 2196–2210.e9 (2018).
Murakami, T. C. et al. A three-dimensional single-cell-resolution whole-brain atlas using CUBIC-X expansion microscopy and tissue clearing. Nat. Neurosci. 21, 625–637 (2018).
Spalteholz, W. Über das Durchsichtigmachen von menschlichen und tierischen Präparaten. (S. Hirzel, 1914).
Erturk, A. et al. Three-dimensional imaging of the unsectioned adult spinal cord to assess axon regeneration and glial responses after injury. Nat. Med. 18, 166–171 (2011).
Erturk, A. et al. Three-dimensional imaging of solvent-cleared organs using 3DISCO. Nat. Protoc. 7, 1983–1995 (2012).
Pan, C. et al. Shrinkage-mediated imaging of entire organs and organisms using uDISCO. Nat. Methods 13, 859–867 (2016).
Chung, K. et al. Structural and molecular interrogation of intact biological systems. Nature 497, 332–337 (2013).
Hogervorst, J. G., Schouten, L. J., Konings, E. J., Goldbohm, R. A. & van den Brandt, P. A. A prospective study of dietary acrylamide intake and the risk of endometrial, ovarian, and breast cancer. Cancer Epidemiol. Biomark. Prev. 16, 2304–2313 (2007).
Tomer, R., Ye, L., Hsueh, B. & Deisseroth, K. Advanced CLARITY for rapid and high-resolution imaging of intact tissues. Nat. Protoc. 9, 1682–1697 (2014).
Yang, B. et al. Single-cell phenotyping within transparent intact tissue through whole-body clearing. Cell 158, 945–958 (2014).
Park, Y.-G. et al. Protection of tissue physicochemical properties using polyfunctional crosslinkers. Nat. Biotechnol. 37, 73 (2018).
Bakutkin, V. V., Maksimova, I. L., Semyonova, T. N., Tuchin, V. V. & Kon., I. L. Controlling of optical properties of sclera. Proc. SPIE 2393, 137–141 (1995).
Tuchin, V. V. et al. Light propagation in tissues with controlled optical properties. J. Biomed. Opt. 2, 401–417 (1997).
Tuchin, V. V. Light scattering of tissues. Physic 40, 495–515 (1997).
Tuchin, V. V. Optical clearing of tissues and blood using the immersion method. J. Phys. D. Appl. Phys. 38, 2497–2518 (2005).
Hama, H. et al. Scale: a chemical approach for fluorescence imaging and reconstruction of transparent mouse brain. Nat. Neurosci. 14, 1481–1488 (2011).
Ke, M. T., Fujimoto, S. & Imai, T. SeeDB: a simple and morphology-preserving optical clearing agent for neuronal circuit reconstruction. Nat. Neurosci. 16, 1154–1161 (2013).
Susaki, E. A. et al. Whole-brain imaging with single-cell resolution using chemical cocktails and computational analysis. Cell 157, 726–739 (2014).
Hama, H. et al. ScaleS: an optical clearing palette for biological imaging. Nat. Neurosci. 18, 1518–1529 (2015).
Perbellini, F. et al. Free-of-acrylamide SDS-based tissue clearing (FASTClear) for three dimensional visualization of myocardial tissue. Sci. Rep. 7, 5188 (2017).
Tainaka, K. et al. Whole-body imaging with single-cell resolution by tissue decolorization. Cell 159, 911–924 (2014).
Liu, Y. C. & Chiang, A. S. High-resolution confocal imaging and three-dimensional rendering. Methods 30, 86–93 (2003).
Helmchen, F. & Denk, W. Deep tissue two-photon microscopy. Nat. Methods 2, 932–940 (2005).
Silvestri, L., Bria, A., Sacconi, L., Iannello, G. & Pavone, F. S. Confocal light sheet microscopy: micron-scale neuroanatomy of the entire mouse brain. Opt. Express 20, 20582–20598 (2012).
Seiriki, K. et al. High-speed and scalable whole-brain imaging in rodents and primates. Neuron 94, 1085–1100.e6 (2017).
Keller, P. J. & Ahrens, M. B. Visualizing whole-brain activity and development at the single-cell level using light-sheet microscopy. Neuron 85, 462–483 (2015).
Helmstaedter, M. & Mitra, P. P. Computational methods and challenges for large-scale circuit mapping. Curr. Opin. Neurobiol. 22, 162–169 (2012).
Ahrens, M. B., Orger, M. B., Robson, D. N., Li, J. M. & Keller, P. J. Whole-brain functional imaging at cellular resolution using light-sheet microscopy. Nat. Methods 10, 413–420 (2013).
Portugues, R., Feierstein, C. E., Engert, F. & Orger, M. B. Whole-brain activity maps reveal stereotyped, distributed networks for visuomotor behavior. Neuron 81, 1328–1343 (2014).
Renier, N. et al. iDISCO: a simple, rapid method to immunolabel large tissue samples for volume imaging. Cell 159, 896–910 (2014).
Amat, F. et al. Efficient processing and analysis of large-scale light-sheet microscopy data. Nat. Protoc. 10, 1679–1696 (2015).
Menegas, W. et al. Dopamine neurons projecting to the posterior striatum form an anatomically distinct subclass. eLife 4, e10032 (2015).
Susaki, E. A. & Ueda, H. R. Whole-body and whole-organ clearing and imaging techniques with single-cell resolution: toward organism-level systems biology in mammals. Cell Chem. Biol. 23, 137–157 (2016).
Tsai, P. S. et al. Correlations of neuronal and microvascular densities in murine cortex revealed by direct counting and colocalization of nuclei and vessels. J. Neurosci. 29, 14553–14570 (2009).
Quan, T. et al. NeuroGPS: automated localization of neurons for brain circuits using L1 minimization model. Sci. Rep. 3, 1414 (2013).
Latorre, A., Alonso-Nanclares, L., Muelas, S., Peña, J.-M. & Defelipe, J. 3D segmentations of neuronal nuclei from confocal microscope image stacks. Front. Neuroanat. 7, 49 (2013).
Frasconi, P. et al. Large-scale automated identification of mouse brain cells in confocal light sheet microscopy images. Bioinformatics 30, i587–i593 (2014).
Amat, F. et al. Fast, accurate reconstruction of cell lineages from large-scale fluorescence microscopy data. Nat. Methods 11, 951–958 (2014).
Wu, J. et al. 3D BrainCV: simultaneous visualization and analysis of cells and capillaries in a whole mouse brain with one-micron voxel resolution. NeuroImage 87, 199–208 (2014).
Quan, T. et al. Digital reconstruction of the cell body in dense neural circuits using a spherical-coordinated variational model. Sci. Rep. 4, 4970 (2014).
Silvestri, L. et al. Quantitative neuroanatomy of all Purkinje cells with light sheet microscopy and high-throughput image analysis. Front. Neuroanat. 9, 68 (2015).
Kim, Y. et al. Brain-wide maps reveal stereotyped cell-type-based cortical architecture and subcortical sexual dimorphism. Cell 171, 456–469.e22 (2017).
Furth, D. et al. An interactive framework for whole-brain maps at cellular resolution. Nat. Neurosci. 21, 139–149 (2018).
Xing, F. & Yang, L. Robust nucleus/cell detection and segmentation in digital pathology and microscopy images: a comprehensive review. IEEE Rev. Biomed. Eng. 9, 234–263 (2016).
Zhang, M., Wu, T. & Bennett, K. M. Small blob identification in medical images using regional features from optimum scale. IEEE Trans. Biomed. Eng. 62, 1051–1062 (2015).
Theer, P., Mongis, C. & Knop, M. PSFj: know your fluorescence microscope. Nat. Methods 11, 981–982 (2014).
Kanda, T., Sullivan, K. F. & Wahl, G. M. Histone-GFP fusion protein enables sensitive analysis of chromosome dynamics in living mammalian cells. Curr. Biol. 8, 377–385 (1998).
Chan, K. Y. et al. Engineered AAVs for efficient noninvasive gene delivery to the central and peripheral nervous systems. Nat. Neurosci. 20, 1172–1179 (2017).
Shinohara, Y. et al. Effects of neutralizing antibody production on AAV-PHP.B-mediated transduction of the mouse central nervous system. Mol. Neurobiol. 56, 4203–4214 (2018).
Sun, Y., Duthaler, S. & Nelson, B. J. Autofocusing in computer microscopy: selecting the optimal focus algorithm. Microsc. Res. Tech. 65, 139–149 (2004).
Royer, L. A. et al. Adaptive light-sheet microscopy for long-term, high-resolution imaging in living organisms. Nat. Biotechnol. 34, 1267–1278 (2016).
Ryan, D. P. et al. Automatic and adaptive heterogeneous refractive index compensation for light-sheet microscopy. Nat. Commun. 8, 612 (2017).
Silvestri, L. et al. RAPID: real-time image-based autofocus for all wide-field optical microscopy systems. Preprint at bioRxiv, https://www.biorxiv.org/content/10.1101/170555v1 (2017).
Royer, L. A., Lemon, W. C., Chhetri, R. K. & Keller, P. J. A practical guide to adaptive light-sheet microscopy. Nat. Protoc. 13, 2462–2500 (2018).
Lindeberg, T. Feature detection with automatic scale selection. Int. J. Comput. Vis. 30, 79–116 (1998).
Hawick, K. A., Leist, A. & Playne, D. P. Parallel graph component labelling with GPUs and CUDA. Parallel Comput. 36, 655–678 (2010).
Soh, Y., Ashraf, H., Hae, Y. & Kim, I. Fast parallel connected component labeling algorithms using CUDA based on 8-directional label selection. Int. J. Latest Res. Sci. Technol. 3, 187–190 (2014).
Frangi, A. F., Niessen, W. J., Vincken, K. L. & Viergever, M. A. Multiscale vessel enhancement filtering. In Medical Image Computing and Computer-Assisted Intervention — MICCAI’98, 130-137 (Springer, 1998).
Avants, B. B. et al. A reproducible evaluation of ANTs similarity metric performance in brain image registration. NeuroImage 54, 2033–2044 (2011).
Pende, M. et al. High-resolution ultramicroscopy of the developing and adult nervous system in optically cleared Drosophila melanogaster. Nat. Commun. 9, 4731 (2018).
Konno, A. & Okazaki, S. Aqueous-based tissue clearing in crustaceans. Zool. Lett. 4, 13 (2018).
Kurihara, D., Mizuta, Y., Sato, Y. & Higashiyama, T. ClearSee: a rapid optical clearing reagent for whole-plant fluorescence imaging. Development 142, 4168–4179 (2015).
Watanave, M. et al. Pharmacological enhancement of retinoid-related orphan receptor alpha function mitigates spinocerebellar ataxia type 3 pathology. Neurobiol. Dis. 121, 263–273 (2019).
Huisken, J. & Stainier, D. Y. Even fluorescence excitation by multidirectional selective plane illumination microscopy (mSPIM). Opt. Lett. 32, 2608–2610 (2007).
Acknowledgements
We thank the lab members at RIKEN BDR and The University of Tokyo, in particular M. Kuroda, for his kind advice on sample gel embedding; D. Shigeta, for his kind help in 3D image preparation; C. Shimizu for support with swelling experiments; and E. A. Susaki, K. Tainaka and A. Kuno for help with developing CUBIC reagents. We thank K. Wilkins for proofreading the manuscript. T.T.M. was supported by the Osaka University Medical Doctor Scientist Training Program. This research was partially supported by the program for Brain Mapping by Integrated Neurotechnologies for Disease Studies (Brain/MINDS) from the Japan Agency for Medical Research and Development, an AMED grant (JP19dm0207057 to H.H.), HFSP (RGP0019/2018 to H.R.U) and JSPS KAKENHI grants 18H02521 (to H.H.) and 19K06899 (to A.K.).
Author information
Authors and Affiliations
Contributions
H.R.U., K.M., T.T.M. and S.A.H. designed the study. T.C.M. developed the CUBIC-X protocols. J.K., T.T.M., T.M. and T.M.W. designed and constructed a customized LSFM. K.M. and T.T.M. developed the MOVIE system. T.M., T.C.M. and S.A.H. prepared the CUBIC-Atlas and analysis programs. H.F. and T.C.M. performed tissue clearing. A.K. and H.H. prepared AAV-PHP.eB (NSE-H2B-mCherry) and the infected mouse. K.M., T.T.M., S.A.H., T.C.M., T.M. and H.R.U. wrote the manuscript. All authors discussed the results and commented on the manuscript text.
Corresponding author
Ethics declarations
Competing interests
H.R.U. and T.C.M. have filed a patent application (PCT/JP2014/070618, 2013-168705) for the CUBIC protocol.
Additional information
Peer review information Nature Protocols thanks Ludovic Silvestri and other anonymous reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key references using this protocol
Murakami, T. C. et al. Nat. Neurosci. 21, 625–637 (2018): https://doi.org/10.1038/s41593-018-0109-1
Tainaka, K. et al. Cell Rep. 24, 2196–2210.e9 (2018): https://doi.org/10.1016/j.celrep.2018.07.056
Kubota, S. I. et al. Cell Rep. 20, 236–250 (2017): https://doi.org/10.1016/j.celrep.2017.06.010
Integrated supplementary information
Supplementary Fig. 1 3D and cross-section images of CUBIC-L/R+ treated brain.
Volume-rendered and cross-section images of CUBIC-L/R+ treated whole mouse brain labelled with H2B-mCherry (green) and RD2 (red). Overlapped signals are shown in yellow. Scale bar, 1mm. All experiments followed governmental and institutional guidelines for the animal experiments.
Supplementary Fig. 2 Parameter optimization for MOVIE-focus.
(a) Optimized six parameters of MOVIE-focus is shown in this table by comprehensive autofocus simulation by using 400,000 images datasets (400 of different light-sheet positions at every 0.35 μm) which was acquired by our LSFM at the 5 μm step size covering 5000 μm depth, using a PI stained mouse brain. Several parts of parameter optimization are shown as an example in (b)-(i). In these panels, only one or two parameters from six parameters are changed. (b)The relationship between block size for image analysis and mean error from ideal focus positions. Error bars show standard deviation of error. (c) The relationship among the block size for image analysis of MOVIE-focus and minimum calculation time for 80% focus accuracy. The focus accuracy was calculated from the ideal focus position, which is determined by the light-sheet position with the highest DCTS score. Although block size 128×128 and 64×64 exhibit similar error in panel (b), the minimum calculation time of block size 128×128 is less than 10 ms per image and faster than that of block size 64×64. (d) The relationship between analyzed area and mean error from ideal focus positions. Error bars show standard deviation of error. (e) The relationship between amplitude of light-sheet oscillation and mean error from ideal focus positions. Error bars show standard deviation of error. (f) The relationship between focus value threshold and mean error from ideal focus positions. Error bars show standard deviation of error. (g) The relationship between focus value threshold and tracking ability. (h) The relationship between block selection order and mean error from ideal focus positions. “Descending” and “Ascending” indicate block selections from top or bottom, respectively. Error bars show standard deviation of error. (i) The relationship between block selection distance (distance function used in block selection; Mean, L1-norm and L2-norm) and mean error from ideal focus positions. Error bars show standard deviation of error.
Supplementary Fig. 3 Result of autofocus simulation for MOVIE performance.
MOVIE-focus was simulated by 400,000 images data sets (400 of different light-sheet positions with 0.35 μm span) which was acquired by our LSFM at the 5 μm step size covering 5000 μm depth, using a PI stained mouse brain. MOVIE (−) used the constant light-sheet position throughout. MOVIE (+) used parameters of Supplementary Fig. 2a. Scale bar, 100 μm.
Supplementary Fig. 4 Result of autofocus simulation for four algorithms.
Result of autofocus simulation. 400,000 images data sets (400 of different light-sheet positions with 0.35 μm span) which was acquired by our LSFM at the 5 μm step size covering 5000 μm depth were used. MOVIE used parameters optimized in Supplementary Fig. 2a. “Without autofocus” means the constant light-sheet position throughout. “Conventional Autofocus” was stop and exposure autofocus mode updated every 1000 μm. An ideal focus position was determined by the light-sheet position with the highest DCTS score of 400 images at each z position. rDCTS (relative DCTS) was calculated with division by DCTS of ideal focus positions. Scale bar, 10 μm.
Supplementary information
Supplementary Information
Supplementary Figures 1–4
Supplementary Video 1. Volume-rendered 3D images of a brain transfected with AAV-PHP.eB: NSE-H2B-mCherry.
Volume-rendered 3D images of a brain transfected with AAV-PHP.eB: NSE-H2B-mCherry (green) counterstained by RD2 (red). The brain is cleared by CUBIC-L/R+ protocol. Overlapped signals are shown in yellow.
Supplementary Video 2. The effect of the MOVIE-focus. Image quality with or without MOVIE-focus.
The left image was acquired with MOVIE-focus, the right was acquired with the constant light-sheet position throughout.
Supplementary Video 3. The reduction of imaging area with MOVIE-skip.
The region surrounded by the yellow frame shows the imaging area without MOVIE-skip. The volume of the image was decreased by more than 50%.
Rights and permissions
About this article
Cite this article
Matsumoto, K., Mitani, T.T., Horiguchi, S.A. et al. Advanced CUBIC tissue clearing for whole-organ cell profiling. Nat Protoc 14, 3506–3537 (2019). https://doi.org/10.1038/s41596-019-0240-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-019-0240-9
This article is cited by
-
Signal improved ultra-fast light-sheet microscope for large tissue imaging
Communications Engineering (2024)
-
Three-dimensional visualization of neural networks inside bone by Osteo-DISCO protocol and alteration of bone remodeling by surgical nerve ablation
Scientific Reports (2023)
-
Three-dimensional mapping in multi-samples with large-scale imaging and multiplexed post staining
Communications Biology (2023)
-
Whole-brain Optical Imaging: A Powerful Tool for Precise Brain Mapping at the Mesoscopic Level
Neuroscience Bulletin (2023)
-
Scalable tissue labeling and clearing of intact human organs
Nature Protocols (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.