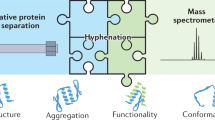
Abstract
Proteins derived by recombinant technologies must be characterized to ensure quality, consistency and optimal production. These properties are usually assayed following purification procedures that are time consuming and labor intensive. Here, we describe a native mass spectrometry (MS) approach, direct-MS, for rapid characterization of intact overexpressed proteins immediately from crude samples. In this protocol, we discuss the multiple applications of the method and outline the necessary steps required for sample preparation, data collection and interpretation of results. We begin with the sample preparation workflows, which are relevant for recombinant proteins produced within bacteria, those analyzed straight from crude cell lysate, and secreted proteins generated in eukaryotic expression systems that are assessed directly from the growth culture medium. We continue with the mass acquisition steps that enable immediate definition of properties such as expressibility, solubility, assembly state, folding, overall structure, stability, post-translational modifications and associations with biomolecules. We demonstrate the applicability of the method by presenting the characterization of a computationally designed toxin–antitoxin heterodimer, activity and protein-interaction determination of a regulatory protein and detailed glycosylation analysis of a designed intact antibody. Overall, we describe a simple and rapid protocol that is relevant to both prokaryotic and eukaryotic expression systems and can be carried out on multiple mass spectrometers, such as Orbitrap and quadrupole time-of-flight (QTOF)-based mass spectroscopy platforms, that enable intact protein detection. The procedure takes from 30 min to several hours, from sample collection to data acquisition, depending on the depth of MS analysis.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout














Similar content being viewed by others
Data availability
The datasets generated during the current study are available from the corresponding author upon reasonable request.
References
Adrio, J. L. & Demain, A. L. Microbial enzymes: tools for biotechnological processes. Biomolecules 4, 117–139 (2014).
Carter, P. J. Introduction to current and future protein therapeutics: a protein engineering perspective. Exp. Cell Res. 317, 1261–1269 (2011).
Gainza-Cirauqui, P. & Correia, B. E. Computational protein design-the next generation tool to expand synthetic biology applications. Curr. Opin. Biotechnol. 52, 145–152 (2018).
Assenberg, R., Wan, P. T., Geisse, S. & Mayr, L. M. Advances in recombinant protein expression for use in pharmaceutical research. Curr. Opin. Struct. Biol. 23, 393–402 (2013).
Holtzhauer, M. Basic Methods for the Biochemical Lab (Springer, 2006).
Miles, A. J. & Wallace, B. A. Biophysical Characterization of Proteins in Developing Biopharmaceuticals (Elsevier, 2015).
Kay, L. E. NMR studies of protein structure and dynamics. J. Magn. Reson. 173, 193–207 (2005).
Ben-Nissan, G. et al. Rapid characterization of secreted recombinant proteins by native mass spectrometry. Commun. Biol. 1, 213 (2018).
Cveticanin, J. et al. Estimating interprotein pairwise interaction energies in cell lysates from a single native mass spectrum. Anal. Chem. 90, 10090–10094 (2018).
Gan, J. et al. Native mass spectrometry of recombinant proteins from crude cell lysates. Anal. Chem. 89, 4398–4404 (2017).
Ghaemmaghami, S. & Oas, T. G. Quantitative protein stability measurement in vivo. Nat. Struct. Biol. 8, 879–882 (2001).
Larsen, M. R., Trelle, M. B., Thingholm, T. E. & Jensen, O. N. Analysis of posttranslational modifications of proteins by tandem mass spectrometry. Biotechniques 40, 790–798 (2006).
Eyers, C. E. & Gaskel, S. J. Mass Spectrometry to Identify Posttranslational Function and Types of Posttranslational Modifications (John Wiley & Sons, 2008).
Christensen, D. G. et al. Mechanisms, detection, and relevance of protein acetylation in prokaryotes. mBio 10, e02708-18 (2019).
St-Denis, N. & Gingras, A. C. Mass spectrometric tools for systematic analysis of protein phosphorylation. Prog. Mol. Biol. Transl. Sci. 106, 3–32 (2012).
Zhou, Q. & Qiu, H. the mechanistic impact of N-glycosylation on stability, pharmacokinetics, and immunogenicity of therapeutic proteins. J. Pharm. Sci. 108, 1366–1377 (2019).
Hernandez, H. & Robinson, C. V. Determining the stoichiometry and interactions of macromolecular assemblies from mass spectrometry. Nat. Protoc. 2, 715–726 (2007).
Wu, L. & Han, D. K. Overcoming the dynamic range problem in mass spectrometry-based shotgun proteomics. Expert Rev. Proteom. 3, 611–619 (2006).
Rose, R. J., Damoc, E., Denisov, E., Makarov, A. & Heck, A. J. High-sensitivity Orbitrap mass analysis of intact macromolecular assemblies. Nat. Methods 9, 1084–1086 (2012).
Chernushevich, I. V. & Thomson, B. A. Collisional cooling of large ions in electrospray mass spectrometry. Anal. Chem. 76, 1754–1760 (2004).
Giles, K. et al. Applications of a travelling wave-based radio-frequency-only stacked ring ion guide. Rapid Commun. Mass Spectrom. 18, 2401–2414 (2004).
Rosano, G. L. & Ceccarelli, E. A. Recombinant protein expression in Escherichia coli: advances and challenges. Front. Microbiol. 5, 172 (2014).
Dumont, J., Euwart, D., Mei, B., Estes, S. & Kshirsagar, R. Human cell lines for biopharmaceutical manufacturing: history, status, and future perspectives. Crit. Rev. Biotechnol. 36, 1110–1122 (2016).
Kebarle, P. & Verkerk, U. H. Electrospray: from ions in solution to ions in the gas phase, what we know now. Mass Spectrom. Rev. 28, 898–917 (2009).
Cech, N. B. & Enke, C. G. Practical implications of some recent studies in electrospray ionization fundamentals. Mass Spectrom. Rev. 20, 362–387 (2001).
Menetret, J. F. et al. Ribosome binding of a single copy of the SecY complex: implications for protein translocation. Mol. Cell 28, 1083–1092 (2007).
Flick, T. G., Cassou, C. A., Chang, T. M. & Williams, E. R. Solution additives that desalt protein ions in native mass spectrometry. Anal. Chem. 84, 7511–7517 (2012).
Hashimoto, K. & Panchenko, A. R. Mechanisms of protein oligomerization, the critical role of insertions and deletions in maintaining different oligomeric states. Proc. Natl Acad. Sci. USA 107, 20352–20357 (2010).
Dixit, S. M., Polasky, D. A. & Ruotolo, B. T. Collision induced unfolding of isolated proteins in the gas phase: past, present, and future. Curr. Opin. Chem. Biol. 42, 93–100 (2018).
Ruotolo, B. T., Benesch, J. L. P., Sandercock, A. M., Hyung, S.-J. & Robinson, C. V. Ion mobility–mass spectrometry analysis of large protein complexes. Nat. Protoc. 3, 1139–1152 (2008).
Toby, T. K. et al. A comprehensive pipeline for translational top-down proteomics from a single blood draw. Nat. Protoc. 14, 119–152 (2019).
van de Waterbeemd, M. et al. Dissecting ribosomal particles throughout the kingdoms of life using advanced hybrid mass spectrometry methods. Nat. Commun. 9, 2493 (2018).
Kaltashov, I. A. & Mohimen, A. Estimates of protein surface areas in solution by electrospray ionization mass spectrometry. Anal. Chem. 77, 5370–5379 (2005).
Sokolovski, M. et al. Measuring inter-protein pairwise interaction energies from a single native mass spectrum by double-mutant cycle analysis. Nat. Commun. 8, 212 (2017).
Chorev, D. S. et al. Protein assemblies ejected directly from native membranes yield complexes for mass spectrometry. Science 362, 829–834 (2018).
Ben-Nissan, G. et al. Triple-stage mass spectrometry unravels the heterogeneity of an endogenous protein complex. Anal. Chem. 89, 4708–4715 (2017).
Allison, T. M. et al. Quantifying the stabilizing effects of protein-ligand interactions in the gas phase. Nat. Commun. 6, 8551 (2015).
Marzahn, M. R. et al. Higher-order oligomerization promotes localization of SPOP to liquid nuclear speckles. EMBO J. 35, 1254–1275 (2016).
Xing, G., Zhang, J., Chen, Y. & Zhao, Y. Identification of four novel types of in vitro protein modifications. J. Proteome Res. 7, 4603–4608 (2008).
Francis, G. L. Albumin and mammalian cell culture: implications for biotechnology applications. Cytotechnology 62, 1–16 (2010).
Darfler, F. J. A protein-free medium for the growth of hybridomas and other cells of the immune system. Vitr. Cell Dev. Biol. 26, 769–778 (1990).
Valdés, R., González, M., Geada, D. & Fernández, E. Assessment of a protein-free medium performance in dfferent cell culture vessels using mouse hybridomas to produce monoclonal antibodies. Pharm. Anal. Acta 3, 155 (2012).
Kirshenbaum, N., Michaelevski, I. & Sharon, M. Analyzing large protein complexes by structural mass spectrometry. J. Vis. Exp. 2010, 1954 (2010).
Lossl, P., Snijder, J. & Heck, A. J. Boundaries of mass resolution in native mass spectrometry. J. Am. Soc. Mass Spectrom. 25, 906–917 (2014).
Netzer, R. et al. Ultrahigh specificity in a network of computationally designed protein-interaction pairs. Nat. Commun. 9, 5286 (2018).
Walsh, G. Biopharmaceutical benchmarks 2018. Nat. Biotechnol. 36, 1136–1145 (2018).
Tiller, K. E. & Tessier, P. M. Advances in antibody design. Annu. Rev. Biomed. Eng. 17, 191–216 (2015).
Liu, H. et al. In vitro and in vivo modifications of recombinant and human IgG antibodies. mAbs 6, 1145–1154 (2014).
Qiu, H. et al. Engineering an anti-CD52 antibody for enhanced deamidation stability. mAbs 11, 1266-1275 (2019).
Jung, S. Y. et al. Complications in the assignment of 14 and 28 Da mass shift detected by mass spectrometry as in vivo methylation from endogenous proteins. Anal. Chem. 80, 1721–1729 (2008).
Afjehi-Sadat, L. & Garcia, B. A. Comprehending dynamic protein methylation with mass spectrometry. Curr. Opin. Chem. Biol. 17, 12–19 (2013).
Brown, C. W. et al. Large-scale analysis of post-translational modifications in E. coli under glucose-limiting conditions. BMC Genomics 18, 301 (2017).
Raftery, M. J. Determination of oxidative protein modifications using mass spectrometry. Redox Rep. 19, 140–147 (2014).
Wingfield, P. T. N-terminal methionine processing. Curr. Protoc. Protein Sci. 88, 6.14.11–16.14.13 (2017).
Lorence, A. Recombinant Gene Expression (Humana Press, 2012).
Liu, H. & May, K. Disulfide bond structures of IgG molecules: structural variations, chemical modifications and possible impacts to stability and biological function. mAbs 4, 17–23 (2012).
Maverakis, E. et al. Glycans in the immune system and The Altered Glycan Theory of Autoimmunity: a critical review. J. Autoimmun. 57, 1–13 (2015).
Manneberg, M., Friedlein, A., Kurth, H., Lahm, H. W. & Fountoulakis, M. Structural analysis and localization of the carbohydrate moieties of a soluble human interferon gamma receptor produced in baculovirus-infected insect cells. Protein Sci. 3, 30–38 (1994).
Morelle, W. & Michalski, J. C. Analysis of protein glycosylation by mass spectrometry. Nat. Protoc. 2, 1585–1602 (2007).
Acknowledgements
We thank E. Morag and E. Bayer, for providing us with the pPICK9 plasmid for CBM3a expression, and Y. Peleg, for providing the pPICK9 plasmid for HSA expression. We are grateful to O. Kersonsky and S. Fleishman for providing us with the plasmid for the expression of MBP-PTE. We also thank S. Warszawski, A. Katz, R. Diskin and S. J. Fleishman, for providing us with the growth media containing secreted antibodies, and R. Diskin, H. Cohen-Dvashi, M. Yona and T. Unger, for providing us with the growth media containing the secreted TfR1. We are also grateful for the support of a Starting Grant from the European Research Council (ERC) (Horizon 2020/ERC grant agreement no. 636752) and for Israel Science Foundation (ISF) grant 300/17. M.S. is the Aharon and Ephraim Katzir Memorial Professorial Chair.
Author information
Authors and Affiliations
Contributions
S.V., G.B.-N. and M.S. designed the experiments. S.V. and G.B.-N performed the experiments. S.V., G.B.-N. and M.S. analyzed the data. M.S., G.B.-N. and S.V. wrote and edited the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Protocols thanks Michael Landreh and other anonymous reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key reference using this protocol
Gan, J. et al. Anal. Chem. 89, 4398–4404 (2017): https://doi.org/10.1021/acs.analchem.7b00398
Key data used in this protocol
Ben-Nissan, G. et al. Commun. Biol. 1, 213 (2018): https://doi.org/10.1038/s42003-018-0231-3
Cveticanin, J. et al. Anal. Chem. 90, 10090–10094 (2018): https://doi.org/10.1021/acs.analchem.8b02349
Supplementary information
Rights and permissions
About this article
Cite this article
Vimer, S., Ben-Nissan, G. & Sharon, M. Direct characterization of overproduced proteins by native mass spectrometry. Nat Protoc 15, 236–265 (2020). https://doi.org/10.1038/s41596-019-0233-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-019-0233-8
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.