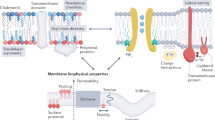
Abstract
Biological membranes define the boundaries of cells and are composed primarily of phospholipids and membrane proteins. It has become increasingly evident that direct interactions of membrane proteins with their surrounding lipids play key roles in regulating both protein conformations and function. However, the exact nature and structural consequences of these interactions remain difficult to track at the molecular level. Here, we present a protocol that specifically addresses this challenge. First, hydrogen–deuterium exchange mass spectrometry (HDX-MS) of membrane proteins incorporated into nanodiscs of controlled lipid composition is used to obtain information on the lipid species that are involved in modulating the conformational changes in the membrane protein. Then molecular dynamics (MD) simulations in lipid bilayers are used to pinpoint likely lipid–protein interactions, which can be tested experimentally using HDX-MS. By bringing together the MD predictions with the conformational readouts from HDX-MS, we have uncovered key lipid–protein interactions implicated in stabilizing important functional conformations. This protocol can be applied to virtually any integral membrane protein amenable to classic biophysical studies and for which a near-atomic-resolution structure or homology model is available. This protocol takes ~4 d to complete, excluding the time for data analysis and MD simulations, which depends on the size of the protein under investigation.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
Data supporting the findings of this article are available from the corresponding author upon reasonable request. All the deuterium uptake plots and uptake datasets of the experiments presented in this work are available on the figshare data repository at https://figshare.com/articles/XylE_HDX-MS_uptake_plots_and_uptake_data/7072988 and https://figshare.com/articles/LacY_HDX-MS_uptake_plots_and_uptake_datasets/7073072
References
Laganowsky, A. et al. Membrane proteins bind lipids selectively to modulate their structure and function. Nature 510, 172–175 (2014).
Martens, C. et al. Direct protein-lipid interactions shape the conformational landscape of secondary transporters. Nat. Commun. 9, 4151 (2018).
Pliotas, C. et al. The role of lipids in mechanosensation. Nat. Struct. Mol. Biol. 22, 991–998 (2015).
Yin, H. & Flynn, A. D. Drugging membrane protein interactions. Annu. Rev. Biomed. Eng. 18, 51–76 (2016).
Gao, Y., Cao, E., Julius, D. & Cheng, Y. TRPV1 structures in nanodiscs reveal mechanisms of ligand and lipid action. Nature 534, 347–351 (2016).
Zhou, M. et al. Mass spectrometry of intact V-type ATPases reveals bound lipids and the effects of nucleotide binding. Science 334, 380–385 (2011).
Allison, T. M. et al. Quantifying the stabilizing effects of protein-ligand interactions in the gas phase. Nat. Commun. 6, 8551 (2015).
Landreh, M. et al. Integrating mass spectrometry with MD simulations reveals the role of lipids in Na(+)/H(+) antiporters. Nat. Commun. 8, 13993 (2017).
Gupta, K. et al. Identifying key membrane protein lipid interactions using mass spectrometry. Nat. Protoc. 13, 1106–1120 (2018).
Pyle, E. et al. Structural lipids enable the formation of functional oligomers of the eukaryotic purine symporter UapA. Cell Chem. Biol. 25, 840–848.e4 (2018).
Sanders, M. R., Findlay, H. E. & Booth, P. J. Lipid bilayer composition modulates the unfolding free energy of a knotted alpha-helical membrane protein. Proc. Natl. Acad. Sci. USA 115, E1799–E1808 (2018).
Alam, A., Kowal, J., Broude, E., Roninson, I. & Locher, K. P. Structural insight into substrate and inhibitor discrimination by human P-glycoprotein. Science 363, 753–756 (2019).
Hunte, C. & Richers, S. Lipids and membrane protein structures. Curr. Opin. Struct. Biol. 18, 406–411 (2008).
Majumdar, D. S. et al. Single-molecule FRET reveals sugar-induced conformational dynamics in LacY. Proc. Natl. Acad. Sci. USA 104, 12640–12645 (2007).
Claxton, D. P., Kazmier, K., Mishra, S. & McHaourab, H. S. Navigating membrane protein structure, dynamics, and energy landscapes using spin labeling and EPR spectroscopy. Methods Enzymol. 564, 349–387 (2015).
Liang, B. & Tamm, L. K. NMR as a tool to investigate the structure, dynamics and function of membrane proteins. Nat. Struct. Mol. Biol. 23, 468–474 (2016).
Konermann, L., Stocks, B. B., Pan, Y. & Tong, X. Mass spectrometry combined with oxidative labeling for exploring protein structure and folding. Mass Spectrom. Rev. 29, 651–667 (2010).
Konijnenberg, A. et al. Global structural changes of an ion channel during its gating are followed by ion mobility mass spectrometry. Proc. Natl. Acad. Sci. USA 111, 17170–17175 (2014).
Marcoux, J. et al. Mass spectrometry reveals synergistic effects of nucleotides, lipids, and drugs binding to a multidrug resistance efflux pump. Proc. Natl. Acad. Sci. USA 110, 9704–9709 (2013).
Vahidi, S., Bi, Y., Dunn, S. D. & Konermann, L. Load-dependent destabilization of the gamma-rotor shaft in FOF1 ATP synthase revealed by hydrogen/deuterium-exchange mass spectrometry. Proc. Natl. Acad. Sci. USA 113, 2412–2417 (2016).
Li, S., Lee, S. Y. & Chung, K. Y. Conformational analysis of g protein-coupled receptor signaling by hydrogen/deuterium exchange mass spectrometry. Methods Enzymol. 557, 261–278 (2015).
Mehmood, S., Domene, C., Forest, E. & Jault, J. M. Dynamics of a bacterial multidrug ABC transporter in the inward- and outward-facing conformations. Proc. Natl. Acad. Sci. USA 109, 10832–10836 (2012).
Zhou, M. et al. Ion mobility-mass spectrometry of a rotary ATPase reveals ATP-induced reduction in conformational flexibility. Nat. Chem. 6, 208–215 (2014).
Eisinger, M. L., Dorrbaum, A. R., Michel, H., Padan, E. & Langer, J. D. Ligand-induced conformational dynamics of the Escherichia coli Na(+)/H(+) antiporter NhaA revealed by hydrogen/deuterium exchange mass spectrometry. Proc. Natl. Acad. Sci. USA 114, 11691–11696 (2017).
Merkle, P. S. et al. Substrate-modulated unwinding of transmembrane helices in the NSS transporter LeuT. Sci. Adv. 4, eaar6179 (2018).
Vadas, O. & Burke, J. E. Probing the dynamic regulation of peripheral membrane proteins using hydrogen deuterium exchange-MS (HDX-MS). Biochem. Soc. Trans. 43, 773–786 (2015).
Harrison, R. A. & Engen, J. R. Conformational insight into multi-protein signaling assemblies by hydrogen-deuterium exchange mass spectrometry. Curr. Opin. Struct. Biol. 41, 187–193 (2016).
Trabjerg, E., Nazari, Z. E. & Rand, K. D. Conformational analysis of complex protein states by hydrogen/deuterium exchange mass spectrometry (HDX-MS): challenges and emerging solutions. Trends Anal. Chem. 106, 125–138 (2018).
Reading, E. et al. Interrogating membrane protein conformational dynamics within native lipid compositions. Angew. Chem. 56, 15654–15657 (2017).
Hebling, C. M. et al. Conformational analysis of membrane proteins in phospholipid bilayer nanodiscs by hydrogen exchange mass spectrometry. Anal. Chem. 82, 5415–5419 (2010).
Adhikary, S. et al. Conformational dynamics of a neurotransmitter:sodium symporter in a lipid bilayer. Proc. Natl. Acad. Sci. USA 114, E1786–E1795 (2017).
Duc, N. M. et al. Effective application of bicelles for conformational analysis of G protein-coupled receptors by hydrogen/deuterium exchange mass spectrometry. J. Am. Soc. Mass Spectrom. 26, 808–817 (2015).
Schuler, M. A., Denisov, I. G. & Sligar, S. G. Nanodiscs as a new tool to examine lipid-protein interactions. Methods Mol. Biol. 974, 415–433 (2013).
Keener, J. E. et al. Chemical additives enable native mass spectrometry measurement of membrane protein oligomeric state within intact nanodiscs. J. Am. Chem. Soc. 141, 1054–1061 (2018).
Marty, M. T., Hoi, K. K., Gault, J. & Robinson, C. V. Probing the lipid annular belt by gas-phase dissociation of membrane proteins in nanodiscs. Angew. Chem. 55, 550–554 (2016).
Murcia Rios, A., Vahidi, S., Dunn, S. D. & Konermann, L. Evidence for a partially stalled gamma rotor in F1-ATPase from hydrogen-deuterium exchange experiments and molecular dynamics simulations. J. Am. Chem. Soc. 140, 14860–14869 (2018).
Skinner, J. J. et al. Benchmarking all-atom simulations using hydrogen exchange. Proc. Natl. Acad. Sci. USA 111, 15975–15980 (2014).
Persson, F. & Halle, B. How amide hydrogens exchange in native proteins. Proc. Natl. Acad. Sci. USA 112, 10383–10388 (2015).
Marrink, S. J. et al. Computational modeling of realistic cell membranes. Chem. Rev. 119, 6184–6226 (2019).
Wen, P. C. et al. Microscopic view of lipids and their diverse biological functions. Curr. Opin. Struct. Biol. 51, 177–186 (2018).
Contreras, F. X. et al. Molecular recognition of a single sphingolipid species by a protein’s transmembrane domain. Nature 481, 525–529 (2012).
Zeppelin, T., Ladefoged, L. K., Sinning, S., Periole, X. & Schiott, B. A direct interaction of cholesterol with the dopamine transporter prevents its out-to-inward transition. PLoS Comput. Biol. 14, e1005907 (2018).
Hedger, G. & Sansom, M. S. P. Lipid interaction sites on channels, transporters and receptors: recent insights from molecular dynamics simulations. Biochim. Biophys. Acta 1858, 2390–2400 (2016).
Salas-Estrada, L. A., Leioatts, N., Romo, T. D. & Grossfield, A. Lipids alter rhodopsin function via ligand-like and solvent-like interactions. Biophys. J. 114, 355–367 (2018).
Frauenfeld, J. et al. A saposin-lipoprotein nanoparticle system for membrane proteins. Nat. Methods 13, 345–351 (2016).
Eswar, N. et al. Comparative protein structure modeling using Modeller. Curr. Protoc. Bioinform. 15, 5.6.1–5.6.30 (2006).
Sun, L. et al. Crystal structure of a bacterial homologue of glucose transporters GLUT1-4. Nature 490, 361–366 (2012).
Abramson, J. et al. Structure and mechanism of the lactose permease of Escherichia coli. Science 301, 610–615 (2003).
West, G. M. et al. Ligand-dependent perturbation of the conformational ensemble for the GPCR beta2 adrenergic receptor revealed by HDX. Structure 19, 1424–1432 (2011).
Yellen, G. The voltage-gated potassium channels and their relatives. Nature 419, 35–42 (2002).
Gurevich, V. V. & Gurevich, E. V. Molecular mechanisms of GPCR signaling: a structural perspective. Int. J. Mol. Sci. 18, E2519 (2017).
Drew, D. & Boudker, O. Shared molecular mechanisms of membrane transporters. Annu. Rev. Biochem. 85, 543–572 (2016).
Denisov, I. G. & Sligar, S. G. Nanodiscs in membrane biochemistry and biophysics. Chem. Rev. 117, 4669–4713 (2017).
Lau, A. M. C., Ahdash, Z., Martens, C. & Politis, A. Deuteros: software for rapid analysis and visualization of data from differential hydrogen deuterium exchange-mass spectrometry. Bioinformatics 35, 3171–3173 (2019).
Manna, M. et al. Mechanism of allosteric regulation of beta2-adrenergic receptor by cholesterol. eLife 5, e18432 (2016).
Guixa-Gonzalez, R. et al. Membrane cholesterol access into a G-protein-coupled receptor. Nat. Commun. 8, 14505 (2017).
Klauda, J. B. et al. Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. J. Phys. Chem. B 114, 7830–7843 (2010).
Phillips, J. C. et al. Scalable molecular dynamics with NAMD. J. Comput. Chem. 26, 1781–1802 (2005).
Humphrey, W., Dalke, A. & Schulten, K. VMD: visual molecular dynamics. J. Mol. Graph. 14, 33–38 (1996).
Jo, S., Kim, T., Iyer, V. G. & Im, W. CHARMM-GUI: a web-based graphical user interface for CHARMM. J. Comput. Chem. 29, 1859–1865 (2008).
Lomize, M. A., Pogozheva, I. D., Joo, H., Mosberg, H. I. & Lomize, A. L. OPM database and PPM web server: resources for positioning of proteins in membranes. Nucleic Acids Res. 40, D370–D376 (2012).
Kandt, C., Ash, W. L. & Tieleman, D. P. Setting up and running molecular dynamics simulations of membrane proteins. Methods 41, 475–488 (2007).
Forest, E. & Man, P. Conformational dynamics and interactions of membrane proteins by hydrogen/deuterium mass spectrometry. Methods Mol. Biol. 1432, 269–279 (2016).
Denisov, I. G. & Sligar, S. G. Nanodiscs for structural and functional studies of membrane proteins. Nat. Struct. Mol. Biol. 23, 481–486 (2016).
Bayburt, T. H. & Sligar, S. G. Membrane protein assembly into nanodiscs. FEBS Lett. 584, 1721–1727 (2010).
Ritchie, T. K. et al. Chapter 11—Reconstitution of membrane proteins in phospholipid bilayer nanodiscs. Methods Enzymol. 464, 211–231 (2009).
Denisov, I. G., Grinkova, Y. V., Lazarides, A. A. & Sligar, S. G. Directed self-assembly of monodisperse phospholipid bilayer nanodiscs with controlled size. J. Am. Chem. Soc. 126, 3477–3487 (2004).
Distler, U., Kuharev, J. & Tenzer, S. Biomedical applications of ion mobility-enhanced data-independent acquisition-based label-free quantitative proteomics. Expert Rev. Proteom. 11, 675–684 (2014).
Martens, C. et al. Lipids modulate the conformational dynamics of a secondary multidrug transporter. Nat. Struct. Mol. Biol. 23, 744–751 (2016).
Sohlenkamp, C. & Geiger, O. Bacterial membrane lipids: diversity in structures and pathways. FEMS Microbiol. Rev. 40, 133–59 (2015).
Acknowledgements
We thank O. Boudker and X. Wang (Weill Cornell Medical College) for providing the GltPh samples. This work was supported by the Wellcome Trust (109854/Z/15/Z) and a King’s Health Partners R&D Challenge Fund through the MRC (MC_PC_15031) to A.P. C.M. was a research fellow from the FRS-FNRS (grant no, 1.B.261.19F). The research was supported in part by the National Institute of General Medical Sciences of the National Institutes of Health under awards U54-GM087519, P41-GM104601 and R01-GM123455 to E.T. The London Interdisciplinary Biosciences Consortium (LIDo) BBSRC Doctoral Training Partnership (BB/M009513/1) supported A.M.L. We also acknowledge computing resources provided by Blue Waters at the National Center for Supercomputing Applications and the Extreme Science and Engineering Discovery Environment (grant TG-MCA06N060 to E.T.).
Author information
Authors and Affiliations
Contributions
C.M. and A.P. designed the experiments. C.M. performed the mutagenesis, expression, purification, reconstitution and HDX-MS experiments. M.S. and E.T. performed and analyzed the MD simulations. C.M. and A.M.L. analyzed the HDX-MS data. C.M. and A.P. wrote the paper with input from all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Journal peer review information Nature Protocols thanks Kasper Rand and other anonymous reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related link
Key reference using this protocol
Martens, C. et al. Nat. Commun. 9, 4151 (2018) https://www.nature.com/articles/s41467-018-06704-1
Supplementary information
Rights and permissions
About this article
Cite this article
Martens, C., Shekhar, M., Lau, A.M. et al. Integrating hydrogen–deuterium exchange mass spectrometry with molecular dynamics simulations to probe lipid-modulated conformational changes in membrane proteins. Nat Protoc 14, 3183–3204 (2019). https://doi.org/10.1038/s41596-019-0219-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-019-0219-6
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.