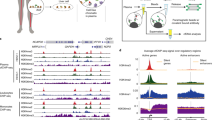
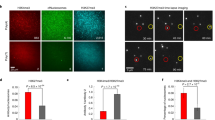
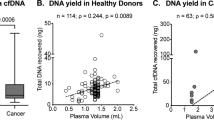
Abstract
Circulating cell-free DNA (cfDNA) comprises small DNA fragments derived from normal and tumor tissue that are released into the bloodstream. Recently, methylation profiling of cfDNA as a liquid biopsy tool has been gaining prominence due to the presence of tissue-specific markers in cfDNA. We have previously reported cell-free methylated DNA immunoprecipitation and high-throughput sequencing (cfMeDIP-seq) as a sensitive, low-input, cost-efficient and bisulfite-free approach to profiling DNA methylomes of plasma cfDNA. cfMeDIP-seq is an extension of a previously published MeDIP-seq protocol and is adapted to allow for methylome profiling of samples with low input (ranging from 1 to 10 ng) of DNA, which is enabled by the addition of ‘filler DNA’ before immunoprecipitation. This protocol is not limited to plasma cfDNA; it can also be applied to other samples that are naturally sheared and at low availability (e.g., urinary cfDNA and cerebrospinal fluid cfDNA), and is potentially applicable to other applications beyond cancer detection, including prenatal diagnostics, cardiology and monitoring of immune response. The protocol presented here should enable any standard molecular laboratory to generate cfMeDIP-seq libraries from plasma cfDNA in ~3–4 d.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
Data from cfMeDIP-seq carried out on a collection of 21 healthy donors were used to generate Fig. 4a,b. Data availability is restricted to processed tables generated from BAM/WIG files due to privacy concerns and to comply with institutional ethics regulations. Processed tables are available at https://github.com/bratmanlab/cfMeDIP_Protocol.
Code availability
The datasets and code are available in the cfMeDIP-seq_Protocol repository at https://github.com/bratmanlab/cfMeDIP_Protocol.
References
Meissner, A. Epigenetic modifications in pluripotent and differentiated cells. Nat. Biotechnol. 28, 1079–1088 (2010).
Varley, K. E. et al. Dynamic DNA methylation across diverse human cell lines and tissues. Genome Res. 23, 555–567 (2013).
Jones, P. A. & Baylin, S. B. The epigenomics of cancer. Cell 128, 683–692 (2007).
Kelly, T. K., De Carvalho, D. D. & Jones, P. A. Epigenetic modifications as therapeutic targets. Nat. Biotechnol. 28, 1069–1078 (2010).
Schwarzenbach, H., Hoon, D. S. B. & Pantel, K. Cell-free nucleic acids as biomarkers in cancer patients. Nat. Rev. Cancer 11, 426–437 (2011).
Corcoran, R. B. & Chabner, B. A. Application of cell-free DNA analysis to cancer treatment. N. Engl. J. Med. 379, 1754–1765 (2018).
Diaz, L. A. & Bardelli, A. Liquid biopsies: genotyping circulating tumor DNA. J. Clin. Oncol. 32, 579–586 (2014).
Wan, J. C. M. et al. Liquid biopsies come of age: towards implementation of circulating tumor DNA. Nat. Rev. Cancer 17, 223–238 (2017).
Newman, A. M. et al. Integrated digital error suppression for improved detection of circulating tumor DNA. Nat. Biotechnol. 34, 547–555 (2016).
Jahr, S. et al. DNA fragments in the blood plasma of cancer patients: quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res. 61, 1659–1665 (2001).
Giacona, M. B. et al. Cell-free DNA in human blood plasma: length measurements in patients with pancreatic cancer and healthy controls. Pancreas 17, 89–97 (1998).
Jung, K., Fleischhacker, M. & Rabien, A. Cell-free DNA in the blood as a solid tumor biomarker—a critical appraisal of the literature. Clin. Chim. Acta 411, 1611–1624 (2010).
Thierry, A. R., El Messaoudi, S., Gahan, P. B., Anker, P. & Stroun, M. Origins, structures, and functions of circulating DNA in oncology. Cancer Metastasis Rev. 35, 347–376 (2016).
Gu, H. et al. Preparation of reduced representation bisulfite sequencing libraries for genome-scale DNA methylation profiling. Nat. Protoc. 6, 468–481 (2011).
Grunau, C., Clark, S. J. & Rosenthal, A. Bisulfite genomic sequencing: systematic investigation of critical experimental parameters. Nucleic Acids Res 29, E65–E65 (2001).
Taiwo, O. et al. Methylome analysis using MeDIP-seq with low DNA concentrations. Nat. Protoc. 7, 617–636 (2012).
Shen, S. Y. et al. Sensitive tumor detection and classification using plasma cell-free DNA methylomes. Nature 563, 579–583 (2018).
Weber, M. et al. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat. Genet. 37, 853–862 (2005).
Yamamoto, N. et al. Detection of aberrant promoter methylation of GSTP1, RASSF1A, and RARβ2 in serum DNA of patients with breast cancer by a newly established one-step methylation-specific PCR assay. Breast Cancer Res. Treat. 132, 165–173 (2012).
Visvanathan, K. et al. Monitoring of serum DNA methylation as an early independent marker of response and survival in metastatic breast cancer: TBCRC 005 prospective biomarker study. J. Clin. Oncol. 35, 751–758 (2016).
Wen, L. et al. Genome-scale detection of hypermethylated CpG islands in circulating cell-free DNA of hepatocellular carcinoma patients. Nat. Publ. Group 25, 1250–1264 (2015).
Irizarry, R. A. et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat. Genet. 41, 178–186 (2009).
Lentini, A. et al. A reassessment of DNA-immunoprecipitation-based genomic profiling. Nat. Methods 15, 499–504 (2018).
Riebler, A. et al. BayMeth: improved DNA methylation quantification for affinity capture sequencing data using a flexible Bayesian approach. Genome Biol. 15, R35 (2014).
Xu, J., Liu, S., Yin, P., Bulun, S. & Dai, Y. MeDEStrand: an improved method to infer genome-wide absolute methylation levels from DNA enrichment data. BMC Bioinformatics 19, 540 (2018).
Lienhard, M. et al. QSEA—modelling of genome-wide DNA methylation from sequencing enrichment experiments. Nucleic Acids Res. 45, e44 (2017).
Zhang, Y. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008).
Fan, H. C., Blumenfeld, Y. J., Chitkara, U., Hudgins, L. & Quake, S. R. Analysis of the size distributions of fetal and maternal cell-free DNA by paired-end sequencing. Clin. Chem. 56, 1279–1286 (2010).
Jiang, P. & Lo, Y. M. D. The long and short of circulating cell-free DNA and the Ins and outs of molecular diagnostics. Trends Genet. 32, 360–371 (2016).
Zhou, Y., Chen, Y., Chen, S. & Gu, J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890 (2018).
Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. Preprint at https://arxiv.org/abs/1303.3997 (2013).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Li, H. et al. The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Lienhard, M., Grimm, C., Morkel, M., Herwig, R. & Chavez, L. MEDIPS: genome-wide differential coverage analysis of sequencing data derived from DNA enrichment experiments. Bioinformatics 30, 284–286 (2013).
Kennedy, S. R. et al. Detecting ultralow-frequency mutations by duplex sequencing. Nat. Protoc. 9, 2586–2606 (2014).
Morgan, M. AnnotationHub: client to access AnnotationHub resources: https://www.bioconductor.org/packages/release/bioc/html/AnnotationHub.html (accessed 1 March 2019).
Acknowledgements
D.D.D.C. and S.V.B. were supported by the Gattuso-Slaight Personalized Cancer Medicine Fund at the Princess Margaret Cancer Centre. J.M.B. was supported by a fellowship from the Strategic Training in Transdisciplinary Radiation Science for the 21st Century (STARS21) training program. D.D.D.C. was supported by the University of Toronto McLaughlin Centre (MC-2015-02); the Canadian Institutes of Health Research (CIHR FDN 148430 and CIHR New Investigator Salary award 201512MSH-360794-228629); the Ontario Institute for Cancer Research (OICR), with funds from the province of Ontario; the Canada Research Chair (950-231346) and the Helen M Cooke Professorship from the Princess Margaret Cancer Foundation. S.V.B. was supported by a Career Development Award from the Conquer Cancer Foundation of ASCO. Any opinions, findings, and conclusions expressed in this article are those of the author(s) and do not necessarily reflect those of the American Society of Clinical Oncology or the Conquer Cancer Foundation. We acknowledge the Princess Margaret Cancer Centre Head & Neck Translational Program, supported by philanthropic funds from the Wharton Family, Joe’s Team, Gordon Tozer and the Reed Fund, as well as the labs of F.-F. Liu and G. Liu (University of Toronto), for the provision of plasma samples. We also thank the Princess Margaret Genomics Centre for carrying out the NGS sequencing and the Bioinformatics and HPC Core of the Princess Margaret Cancer Centre for expertise in generating the NGS data.
Author information
Authors and Affiliations
Contributions
S.Y.S. and D.D.D.C. designed and developed the cfMeDIP-seq protocol. S.Y.S. and J.M.B. performed and optimized the protocol with input from S.V.B. and D.D.D.C. S.Y.S. wrote the manuscript with assistance from J.M.B., S.V.B. and D.D.D.C. J.M.B performed the bioinformatic analysis of all representative cfMeDIP-seq libraries described in the article. All authors contributed to, reviewed and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
D.D.D.C., S.Y.S., S.V.B. and J.M.B are listed as inventors/contributors on patents filed that are related to this work. S.V.B. is a co-inventor on a patent related to mutation-based ctDNA detection technology that has been licensed to Roche Molecular Diagnostics. D.D.D.C. and S.V.B. have received research funding from Nektar Therapeutics.
Additional information
Peer review information: Nature Protocols thanks Peter W. Laird, Eleni Tomazou and other anonymous reviewer(s) for their contribution to the peer review of this work.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Protocol to which this paper is an extension
Taiwo, O. et al. Nat. Protoc. 7, 617–636 (2012) https://doi.org/10.1038/nprot.2012.012
Key reference using this protocol
Shen, S. Y. et al. Nature 563, 579–583 (2018): https://doi.org/10.1038/s41586-018-0703-0
This protocol is an extension to: Nat. Protoc. 7, 617–636 (2012), doi:10.1038/nprot.201.012
Supplementary information
Rights and permissions
About this article
Cite this article
Shen, S.Y., Burgener, J.M., Bratman, S.V. et al. Preparation of cfMeDIP-seq libraries for methylome profiling of plasma cell-free DNA. Nat Protoc 14, 2749–2780 (2019). https://doi.org/10.1038/s41596-019-0202-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-019-0202-2
This article is cited by
-
Liquid biopsy of cerebrospinal fluid enabling the detection and therapeutic targeting of the BRAFV600E mutation in a patient with overlapping Erdheim-Chester/Rosai-Dorfman disease
Journal of Neurology (2024)
-
Unlocking the secrets: the power of methylation-based cfDNA detection of tissue damage in organ systems
Clinical Epigenetics (2023)
-
Altered cfDNA fragmentation profile in hypomethylated regions as diagnostic markers in breast cancer
Epigenetics & Chromatin (2023)
-
Early detection of lung cancer using artificial intelligence-enhanced optical nanosensing of chromatin alterations in field carcinogenesis
Scientific Reports (2023)
-
The methylome and cell-free DNA: current applications in medicine and pediatric disease
Pediatric Research (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.