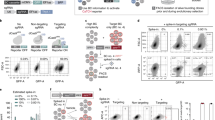
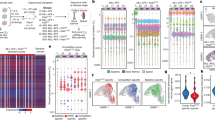
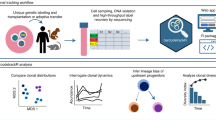
Abstract
Lineage tracing is a powerful tool that can be used to uncover cell fates. Here, we describe a novel method for the quantitative analysis of clonal dynamics in grafted cancer tissues. The protocol involves the preparation and validation of cells for lineage tracing, establishment of grafts and label induction, analysis of clone-size distribution and fitting of the experimental data to a mathematical tumor growth model. In contrast to other lineage-tracing strategies, the method described here assesses stem cell functionality and infers tumor expansion dynamics independently of molecular markers such as putative cancer stem cell (CSC)-specific genes. The experimental system and analytical framework presented can be used to quantify clonal advantages that specific mutations provide, in both the absence and presence of (targeted) therapeutic agents. This protocol typically takes ~20 weeks to complete from cell line selection to inference of growth dynamics, depending on the grafted cancer growth rate.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout











Similar content being viewed by others
Data availability
Source data for Figs. 7, 8, 9 and 10 have been provided as Supplementary Data. All other data supporting the findings of this study are available from the corresponding authors on reasonable request.
Code availability
The custom clone-size quantification software can be downloaded from https://github.com/dmmiedema/Clone-Sizes-From-Image. The CloneFit app can be downloaded from https://github.com/dmmiedema/CloneFit. We have made available the programming code for the tumor growth model on GitHub: https://github.com/dmmiedema/Tumor-Growth-Model.
References
Todaro, M. et al. Colon cancer stem cells dictate tumor growth and resist cell death by production of interleukin-4. Cell Stem Cell 1, 389–402 (2007).
O’Brien, C. A., Pollett, A., Gallinger, S. & Dick, J. E. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature 445, 106–110 (2007).
Ricci-Vitiani, L. et al. Identification and expansion of human colon-cancer-initiating cells. Nature 445, 111–115 (2007).
Lenos, K. J. et al. Stem cell functionality is microenvironmentally defined during tumour expansion and therapy response in colon cancer. Nat. Cell Biol. 20, 1193–1202 (2018).
Vermeulen, L. & Snippert, H. J. Stem cell dynamics in homeostasis and cancer of the intestine. Nat. Rev. Cancer 14, 468–480 (2014).
Shimokawa, M. et al. Visualization and targeting of LGR5+ human colon cancer stem cells. Nature 545, 187–192 (2017).
De Sousa e Melo, F. et al. A distinct role for Lgr5+ stem cells in primary and metastatic colon cancer. Nature 543, 676–680 (2017).
Flanagan, D. J., Hodder, M. C. & Sansom, O. J. Microenvironmental cues in cancer stemness. Nat. Cell Biol. 20, 1102–1104 (2018).
Rodriguez, E. et al. Versatile and enhanced tumour modelling in mice via somatic cell transduction. J. Pathol. 232, 449–457 (2014).
Vermeulen, L. et al. Defining stem cell dynamics in models of intestinal tumor initiation. Science 342, 995–998 (2013).
Mascre, G. et al. Distinct contribution of stem and progenitor cells to epidermal maintenance. Nature 489, 257–262 (2012).
Rulands, S. et al. Universality of clone dynamics during tissue development. Nat. Phys. 14, 469–474 (2018).
Zhang, Z. & Lutz, B. Cre recombinase-mediated inversion using lox66 and lox71: method to introduce conditional point mutations into the CREB-binding protein. Nucleic Acids Res. 30, e90 (2002).
Fumagalli, A. et al. A surgical orthotopic organoid transplantation approach in mice to visualize and study colorectal cancer progression. Nat. Protoc. 13, 235–247 (2018).
Roper, J. et al. Colonoscopy-based colorectal cancer modeling in mice with CRISPR-Cas9 genome editing and organoid transplantation. Nat. Protoc. 13, 217–234 (2018).
Pavia-Jimenez, A., Tcheuyap, V. T. & Brugarolas, J. Establishing a human renal cell carcinoma tumorgraft platform for preclinical drug testing. Nat. Protoc. 9, 1848–1859 (2014).
Lorenzatti Hiles, G. et al. A surgical orthotopic approach for studying the invasive progression of human bladder cancer. Nat. Protoc. 14, 738–755 (2019).
Kim, M. P. et al. Generation of orthotopic and heterotopic human pancreatic cancer xenografts in immunodeficient mice. Nat. Protoc. 4, 1670–1680 (2009).
Huynh, H., Soo, K. C., Chow, P. K., Panasci, L. & Tran, E. Xenografts of human hepatocellular carcinoma: a useful model for testing drugs. Clin. Cancer Res. 12, 4306–4314 (2006).
van der Heijden, M. et al. Spatiotemporal regulation of clonogenicity in colorectal cancer xenografts. Proc. Natl. Acad. Sci. USA 116, 6140–6145 (2019).
Driessens, G., Beck, B., Caauwe, A., Simons, B. D. & Blanpain, C. Defining the mode of tumour growth by clonal analysis. Nature 488, 527–530 (2012).
Frick, P. L., Paudel, B. B., Tyson, D. R. & Quaranta, V. Quantifying heterogeneity and dynamics of clonal fitness in response to perturbation. J. Cell. Physiol. 230, 1403–1412 (2015).
Gillespie, D. T. Exact stochastic simulation of coupled chemical reactions. J. Phys. Chem. 81, 2340–2361 (1977).
Acknowledgements
We thank L.E. Nijman and M.C. Lecca for their help with the animal procedures. This work was supported by the Academic Medical Center (Amsterdam), The New York Stem Cell Foundation, Cancer Research UK, and grants from KWF (UVA2011-4969, UVA2014-7245 and 10529), the Maurits en Anna de Kock Stichting (2015-2), Worldwide Cancer Research (14-1164), the Maag Lever Darm Stichting (MLDS-CDG 14-03), the European Research Council (ERC-StG 638193) and ZonMw (Vidi 016.156.308) to L.V. L.V. is a New York Stem Cell Foundation—Robertson Investigator.
Author information
Authors and Affiliations
Contributions
K.J.L. and S.C.L. performed the experiments; D.M.M. and L.V. developed the quantitative models; D.M.M. developed the software; K.J.L., D.M.M., S.C.L., M.F.B. and L.V. analyzed the data; S.K.L. contributed reagents; K.J.L., D.M.M., M.F.B. and L.V. conceived and designed the research; K.J.L., S.C.L., D.M.M., M.F.B. and L.V. wrote the manuscript; L.V. directed the research. All authors approved the content of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key reference using this protocol
Lenos, K. J. et al. Nat. Cell Biol. 20, 1193–1202 (2018): https://doi.org/10.1038/s41556-018-0179-z
Supplementary information
Supplementary Data
HT55 clone size data
Rights and permissions
About this article
Cite this article
Lenos, K.J., Lodestijn, S.C., Lyons, S.K. et al. A marker-independent lineage-tracing system to quantify clonal dynamics and stem cell functionality in cancer tissue. Nat Protoc 14, 2648–2671 (2019). https://doi.org/10.1038/s41596-019-0194-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-019-0194-y
This article is cited by
-
Emerging role of G9a in cancer stemness and promises as a therapeutic target
Oncogenesis (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.