Abstract
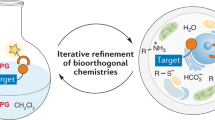
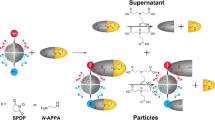
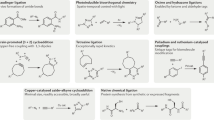
A requirement for biochemical labeling strategies is a pronounced biocompatibility of the underlying reaction methodology. This protocol enables a systematic evaluation of the biocompatibility of (new) reaction methodologies that are potentially attractive for biochemical applications. The cellular environment for in vitro and in vivo applications is mimicked by the one-by-one addition of diverse bio-additives to the reaction. The influence of the bio-additives on the product yield, termed bio-robustness, is quantified by gas chromatography (GC) or NMR techniques, whereas qualitative analysis of the level of biomolecule preservation by ultra-HPLC–mass spectrometry (UHPLC–MS) or gel electrophoresis enables monitoring of the effects of the reaction conditions on the biomolecule stability, e.g., bio-additive modification or degradation. The 22 chosen bio-additives and the required controls can be completely evaluated within 5–7 working days, depending on reaction time, instrument and the general equipment availability of the lab. We illustrate this protocol by assessing the reaction biocompatibility of a copper-catalyzed N-arylation of sulfonamides. The hereby obtained results are compared to those for a reaction that is characterized by high reaction biocompatibility: the energy-transfer-enabled disulfide–ene reaction.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
The data supporting the findings of this study are available within the paper and its supplementary information, or from the corresponding authors upon reasonable request.
References
Trost, B. M. Selectivity: a key to synthetic efficiency. Science 219, 245–250 (1983).
Wender, P. A. & Miller, B. L. Synthesis at the molecular frontier. Nature 460, 197–201 (2009).
Sletten, E. M. & Bertozzi, C. R. Bioorthogonal chemistry: fishing for selectivity in a sea of functionality. Angew. Chem. Int. Ed. 48, 6974–6998 (2009).
Devaraj, N. K. The future of bioorthogonal chemistry. ACS Cent. Sci. 4, 952–959 (2018).
Vert, M. et al. Terminology for biorelated polymers and applications (IUPAC Recommendations 2012). Pure Appl. Chem. 84, 377–410 (2012).
Definition of biocompatibility. https://www.merriam-webster.com/dictionary/biocompatibility (2018).
Marks, K. M. & Nolan, G. P. Chemical labeling strategies for cell biology. Nat. Methods 3, 591–596 (2006).
Yang, M., Li, J. & Chen, P. R. Transition metal-mediated bioorthogonal protein chemistry in living cells. Chem. Soc. Rev. 43, 6511–6526 (2014).
Christoffel, F. & Ward, T. R. Palladium-catalyzed heck cross-coupling reactions in water: a comprehensive review. Catal. Lett. 148, 489–511 (2018).
Tsubokura, K. et al. In vivo gold complex catalysis within live mice. Angew. Chem. Int. Ed. 56, 3579–3584 (2017).
Ngo, A. H., Bose, S. & Do, L. H. Intracellular chemistry: integrating molecular inorganic catalysts with living systems. Chem. Eur. J 24, 10584–10594 (2018).
Chandrasekaran, K. S. & Rentmeister, A. Clicking a fish: click chemistry of different biomolecules in Danio rerio. Biochemistry, 58, 24–30.
Williams, D. F. On the mechanisms of biocompatibility. Biomaterials 29, 2941–2953 (2008).
Collins, K. D. & Glorius, F. A robustness screen for the rapid assessment of chemical reactions. Nat. Chem 5, 597–601 (2013).
Collins, K. D., Rühling, A. & Glorius, F. Application of a robustness screen for the evaluation of synthetic organic methodology. Nat. Protoc. 9, 1348–1353 (2014).
Gensch, T., Teders, M. & Glorius, F. Approach to comparing the functional group tolerance of reactions. J. Org. Chem. 82, 9154–9159 (2017).
Collins, K. D. & Glorius, F. Intermolecular reaction screening as a tool for reaction evaluation. Acc. Chem. Res. 48, 619–627 (2015).
Richardson, J., Ruble, J. C., Love, E. A. & Berritt, S. A method for identifying and developing functional group tolerant catalytic reactions: application to the Buchwald–Hartwig amination. J. Org. Chem. 82, 3741–3750 (2017).
Lin, S. et al. Mapping the dark space of chemical reactions with extended nanomole synthesis and MALDI-TOF MS. Science 361, eaar6236 (2018).
Teders, M. et al. The energy transfer enabled biocompatible disulfide–ene reaction. Nat. Chem. 10, 981–988 (2018).
Forouhar, F. et al. Two Fe-S cluster catalyse sulfur insertion by radical-SAM methylthiotransferases. Nat. Chem. Biol. 9, 333–338 (2013).
Chen, Y., Kamlet, A. S., Steinman, J. B. & Liu, D. R. A biomolecule-compatible visible-light-induced azide reduction from a DNA-encoded reaction-discovery system. Nat. Chem 3, 146–153 (2011).
Huang, H., Zhang, G., Gong, L., Zhang, S. & Chen, Y. Visible-light-induced chemoselective deboronative alkynylation under biomolecule-compatible conditions. J. Am. Chem. Soc. 136, 2280–2283 (2014).
Zhu, Y., Bauer, M. & Ackermann, L. Late-stage peptide diversification by bioorthogonal catalytic C–H arylation at 23 °C in H2O. Chem. Eur. J 21, 9980–9983 (2015).
Good, N. E. et al. Ion buffers for biological research. Biochemistry 5, 467–477 (1966).
Good, N. E. & Izawa, S. Hydrogen ion buffers. Methods Enzymol 24, 53–68 (1972).
Ferguson, W. J. et al. Hydrogen ion buffers for biological research. Anal. Biochem. 104, 300–310 (1980).
Gong, Y. & Pan, L. Recent advances in bioorthogonal reactions for site-specific protein labeling and engineering. Tetrahedron Lett. 56, 2123–2132 (2015).
Bennett, B. D. et al. Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nat. Chem. Biol. 5, 593–599 (2009).
Giustarini, D. et al. Glutathione, glutathionedisulfide, and S-glutathionylatedproteins in cell cultures. Free Radic. Biol. Med. 89, 971–981 (2015).
Geng, X. et al. Copper-catalyzed direct N-arlyation of N-arylsulfonanilides using diaryliodonium salts in water. Tetrahedron Lett. 55, 3856–3859 (2014).
Linder, M. C. The relationship of copper to DNA damage and damage prevention in humans. Mutat. Res. 733, 83–91 (2012).
Acknowledgements
We thank S. Hüwel, W. Dörner and S. Wulff for experimental and technical assistance (all WWU Münster). This work was supported by the Deutsche Forschungsgemeinschaft (Leibniz Award to F.G. and RE2796/6-1 to A.R.) and by the Fonds der Chemischen Industrie (doctoral fellowship to L.A. and Dozentenpreis to A.R.). M.T. thanks SusChemSys 2.0 for general support.
Author information
Authors and Affiliations
Contributions
L.A., M.T., A.R. and F.G. designed the concept and the protocol. L.A. and M.T. performed all experimental work. All authors co-wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information: Nature Protocols thanks Gonçalo Bernardes and other anonymous reviewer(s) for their contribution to the peer review of this work.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key reference using this protocol
Teders, M. et al. Nat. Chem. 10, 981–988 (2018): https://www.nature.com/articles/s41557-018-0102-z
Supplementary information
Supplementary Information
Supplementary Figures 1–23, Supplementary Table 1, Supplementary Note 1 and Supplementary Procedure 1
Rights and permissions
About this article
Cite this article
Anhäuser, L., Teders, M., Rentmeister, A. et al. Bio-additive-based screening: toward evaluation of the biocompatibility of chemical reactions. Nat Protoc 14, 2599–2626 (2019). https://doi.org/10.1038/s41596-019-0190-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-019-0190-2
This article is cited by
-
Bridging the information gap in organic chemical reactions
Nature Chemistry (2024)
-
Electrochemical gold-catalysed biocompatible C(sp2)–C(sp) coupling
Nature Synthesis (2023)
-
Flow parallel synthesizer for multiplex synthesis of aryl diazonium libraries via efficient parameter screening
Communications Chemistry (2021)
-
Design and evolution of chimeric streptavidin for protein-enabled dual gold catalysis
Nature Catalysis (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.