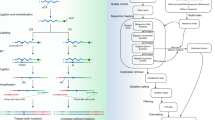
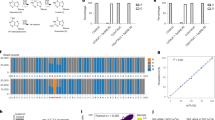
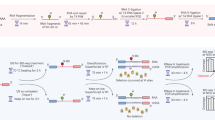
Abstract
Advances in RNA-sequencing methods have uncovered many aspects of RNA metabolism but are limited to surveying either the 3ʹ or 5ʹ terminus of RNAs, thus missing mechanistic aspects that could be revealed if both ends were captured. We developed Akron sequencing (Akron-seq), a method that captures in parallel the native 5ʹ ends of uncapped, polyadenylated mRNAs and 3ʹ ends of capped mRNAs from the same input RNA. Thus, Akron-seq uniquely enables assessment of full-length and truncated mRNAs at single-nucleotide resolution. Akron-seq involves RNA isolation, depletion of ribosomal and abundant small capped RNAs, and selection of capped and polyadenylated mRNAs. The endogenous ends of mRNAs are marked by adaptor ligation, followed by fragmentation, cDNA generation, PCR amplification, and deep sequencing. The step-by-step protocol we describe here is optimized for cultured human cells but can be adapted to primary cells and tissues. Akron-seq can be completed within 6 d, and sequencing and analysis can be completed within 6 d.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
Previous published datasets generated or analyzed using the current protocol are available in the Gene Expression Omnibus repository, under accession no. GSE107838.
Code availability
Previous published source code required for analysis has been deposited on GitHub (https://github.com/mnsmar/ribothrypsis).
References
Green, R. & Noller, H. F. Ribosomes and translation. Annu. Rev. Biochem. 66, 679–716 (1997).
Coller, J. & Parker, R. Eukaryotic mRNA decapping. Annu. Rev. Biochem. 73, 861–890 (2004).
Ghosh, S. & Jacobson, A. RNA decay modulates gene expression and controls its fidelity. Wiley Interdiscip. Rev. RNA 1, 351–361 (2010).
Parker, R. RNA degradation in Saccharomyces cerevisae. Genetics 191, 671–702 (2012).
Anderson, J. S. & Parker, R. P. The 3ʹ to 5ʹ degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SKI2 DEVH box protein and 3ʹ to 5ʹ exonucleases of the exosome complex. EMBO J. 17, 1497–1506 (1998).
Schoenberg, D. R. & Maquat, L. E. Regulation of cytoplasmic mRNA decay. Nat. Rev. Genet. 13, 246–259 (2012).
Garneau, N. L., Wilusz, J. & Wilusz, C. J. The highways and byways of mRNA decay. Nat. Rev. Mol. Cell. Biol. 8, 113–126 (2007).
Lykke-Andersen, S., Tomecki, R., Jensen, T. H. & Dziembowski, A. The eukaryotic RNA exosome: same scaffold but variable catalytic subunits. RNA Biol. 8, 61–66 (2011).
Frischmeyer, P. A. et al. An mRNA surveillance mechanism that eliminates transcripts lacking termination codons. Science 295, 2258–2261 (2002).
Doma, M. K. & Parker, R. Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature 440, 561–564 (2006).
Isken, O. & Maquat, L. E. Quality control of eukaryotic mRNA: safeguarding cells from abnormal mRNA function. Genes Dev. 21, 1833–1856 (2007).
Shoemaker, C. J. & Green, R. Translation drives mRNA quality control. Nat. Struct. Mol. Biol. 19, 594–601 (2012).
Mendell, J. T., Sharifi, N. A., Meyers, J. L., Martinez-Murillo, F. & Dietz, H. C. Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nat. Genet. 36, 1073–1078 (2004).
Guydosh, N. R. & Green, R. Translation of poly(A) tails leads to precise mRNA cleavage. RNA 23, 749–761 (2017).
Lykke-Andersen, S. & Jensen, T. H. Nonsense-mediated mRNA decay: an intricate machinery that shapes transcriptomes. Nat. Rev. Mol. Cell Biol. 16, 665–677 (2015).
Pelechano, V., Wei, W. & Steinmetz, L. M. Widespread co-translational RNA decay reveals ribosome dynamics. Cell 161, 1400–1412 (2015).
Hu, W., Sweet, T. J., Chamnongpol, S., Baker, K. E. & Coller, J. Co-translational mRNA decay in Saccharomyces cerevisiae. Nature 461, 225–229 (2009).
Yu, X., Willmann, M. R., Anderson, S. J. & Gregory, B. D. Genome-wide mapping of uncapped and cleaved transcripts reveals a role for the nuclear mRNA cap-binding complex in cotranslational RNA decay in Arabidopsis. Plant Cell 28, 2385–2397 (2016).
Ibrahim, F., Maragkakis, M., Alexiou, P. & Mourelatos, Z. Ribothrypsis, a novel process of canonical mRNA decay, mediates ribosome-phased mRNA endonucleolysis. Nat. Struct. Mol. Biol. 25, 302–310 (2018).
Ingolia, N. T., Ghaemmaghami, S., Newman, J. R. & Weissman, J. S. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324, 218–223 (2009).
Rouskin, S., Zubradt, M., Washietl, S., Kellis, M. & Weissman, J. S. Genome-wide probing of RNA structure reveals active unfolding of mRNA structures in vivo. Nature 505, 701–705 (2014).
Kwok, C. K., Marsico, G., Sahakyan, A. B., Chambers, V. S. & Balasubramanian, S. rG4-seq reveals widespread formation of G-quadruplex structures in the human transcriptome. Nat. Methods 13, 841–844 (2016).
Lubas, M. et al. The human nuclear exosome targeting complex is loaded onto newly synthesized RNA to direct early ribonucleolysis. Cell Rep. 10, 178–192 (2015).
Carlevaro-Fita, J., Rahim, A., Guigo, R., Vardy, L. A. & Johnson, R. Cytoplasmic long noncoding RNAs are frequently bound to and degraded at ribosomes in human cells. RNA 22, 867–882 (2016).
Addo-Quaye, C., Eshoo, T. W., Bartel, D. P. & Axtell, M. J. Endogenous siRNA and miRNA targets identified by sequencing of the Arabidopsis degradome. Curr. Biol. 18, 758–762 (2008).
German, M. A. et al. Global identification of microRNA-target RNA pairs by parallel analysis of RNA ends. Nat. Biotechnol. 26, 941–946 (2008).
Schmidt, S. A. et al. Identification of SMG6 cleavage sites and a preferred RNA cleavage motif by global analysis of endogenous NMD targets in human cells. Nucleic Acids Res. 43, 309–323 (2015).
Chang, H., Lim, J., Ha, M. & Kim, V. N. TAIL-seq: genome-wide determination of poly(A) tail length and 3ʹ end modifications. Mol. Cell 53, 1044–1052 (2014).
Subtelny, A. O., Eichhorn, S. W., Chen, G. R., Sive, H. & Bartel, D. P. Poly(A)-tail profiling reveals an embryonic switch in translational control. Nature 508, 66–71 (2014).
Tarailo-Graovac, M. & Chen, N. Using RepeatMasker to identify repetitive elements in genomic sequences. Curr. Protoc. Bioinformatics Chapter 4, Unit 4.10 (2009).
Maragkakis, M., Alexiou, P., Nakaya, T. & Mourelatos, Z. CLIPSeqTools—a novel bioinformatics CLIP-seq analysis suite. RNA 22, 1–9 (2015).
Martin, M. CUTADAPT removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 10–12 (2011).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2012).
Acknowledgements
We thank the members of the laboratory of Z.M. and T. Jaber for helpful discussions. We thank M. Maragkakis for help in designing custom snRNAs oligonucleotides. This study was technically supported by the Functional Genomics Core at the University of Pennsylvania. This study was financially supported by grants from the ALS Therapy Alliance (2013-S-014) and the NIH (GM072777) to Z.M.
Author information
Authors and Affiliations
Contributions
F.I. and Z.M. conceived Akron-seq protocol. F.I. developed, optimized, performed, and interpreted the Akron-seq results with insightful input from Z.M. F.I. wrote the manuscript with input and editing from Z.M.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Journal peer review information: Nature Protocols thanks Kristian Baker and other anonymous reviewer(s) for their contribution to the peer review of this work.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related link
Key reference using this protocol
Ibrahim, F., Maragkakis, M., Alexiou, P. & Mourelatos, Z. Nat. Struct. Mol. Biol. 25, 302–310 (2018): https://doi.org/10.1038/s41594-018-0042-8
Supplementary information
Rights and permissions
About this article
Cite this article
Ibrahim, F., Mourelatos, Z. Capturing 5ʹ and 3ʹ native ends of mRNAs concurrently with Akron sequencing. Nat Protoc 14, 1578–1602 (2019). https://doi.org/10.1038/s41596-019-0151-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-019-0151-9
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.