Abstract
Non-coding RNA (ncRNA) molecules have been shown to play a variety of cellular roles; however, the contributions of different types of RNA to specific phenomena are often hard to dissect. To study the role of RNA in the assembly of DNA damage response (DDR) foci, we developed the RNase A treatment and reconstitution (RATaR) method, in which cells are mildly permeabilized, incubated with recombinant RNase A and subsequently reconstituted with different RNA species, under conditions of RNase A inactivation and inhibition of endogenous transcription. The block of transcription right after RNase A removal represents a key innovation of RATaR, preventing potential contributions of endogenously neo-synthesized transcripts to the phenotypes studied. A critical aspect of this technique is the balance between sufficient permeabilization of membranes to allow enzyme/RNA access into the cell nucleus and cell viability. Here, we present our protocol for RNA-dependent DDR foci disassembly and reassembly using fluorescent DDR RNAs (DDRNAs) in NIH2/4 cells, an engineered NIH3T3-derived cell line. The use of sequence-specific, fluorescent RNA molecules permits the concomitant determination of their subcellular localization and biological functions. We also outline adaptations of RATaR when implemented in different cell lines exposed to various genotoxic treatments, such as γ-radiation, restriction enzymes and telomere deprotection. In all these cases, the entire procedure can be completed within 2 h without the need for special equipment or uncommon skills. We believe this technique will prove useful for investigating the contribution of RNA to a variety of relevant cellular processes.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Michelini, F. et al. From “cellular” RNA to “smart” RNA: multiple roles of RNA in genome stability and beyond. Chem. Rev. 118, 4365–4403 (2018).
Francia, S. Non-coding RNA: sequence-specific guide for chromatin modification and DNA damage signaling. Front. Genet. 6, 320 (2015).
Francia, S. et al. Site-specific DICER and DROSHA RNA products control the DNA-damage response. Nature 488, 231–235 (2012).
Maison, C. et al. Higher-order structure in pericentric heterochromatin involves a distinct pattern of histone modification and an RNA component. Nat. Genet. 30, 329–334 (2002).
Pryde, F. et al. 53BP1 exchanges slowly at the sites of DNA damage and appears to require RNA for its association with chromatin. J. Cell Sci. 118, 2043–2055 (2005).
Francia, S., Cabrini, M., Matti, V., Oldani, A. & d’Adda di Fagagna, F. DICER, DROSHA and DNA damage response RNAs are necessary for the secondary recruitment of DNA damage response factors. J. Cell Sci. 129, 1468–1476 (2016).
Michelini, F. et al. Damage-induced lncRNAs control the DNA damage response through interaction with DDRNAs at individual double-strand breaks. Nat. Cell Biol. 19, 1400–1411 (2017).
Rossiello, F. et al. DNA damage response inhibition at dysfunctional telomeres by modulation of telomeric DNA damage response RNAs. Nat. Commun. 8, 13980 (2017).
Okamoto, K. et al. A two-step mechanism for TRF2-mediated chromosome-end protection. Nature 494, 502–505 (2013).
Aguzzi, A. & Altmeyer, M. Phase separation: linking cellular compartmentalization to disease. Trends Cell Biol. 26, 547–558 (2016).
Wei, W. et al. A role for small RNAs in DNA double-strand break repair. Cell 149, 101–112 (2012).
Gao, M. et al. Ago2 facilitates Rad51 recruitment and DNA double-strand break repair by homologous recombination. Cell Res. 24, 532–541 (2014).
Wang, Q. & Goldstein, M. Small RNAs recruit chromatin-modifying enzymes MMSET and Tip60 to reconfigure damaged DNA upon double-strand break and facilitate repair. Cancer Res. 76, 1904–1915 (2016).
Patne, K. et al. BRG1 and SMARCAL1 transcriptionally co-regulate DROSHA, DGCR8 and DICER in response to doxorubicin-induced DNA damage. Biochim. Biophys. Acta Gene Regul. Mech. 1860, 936–951 (2017).
Cold Spring Harbor Laboratory Press. Cytoskeletal (CSK) buffer. Cold Spring Harb. Protoc. https://doi.org/10.1101/pdb.rec084301 (2016).
Amidzadeh, Z. et al. Assessment of different permeabilization methods of minimizing damage to the adherent cells for detection of intracellular RNA by flow cytometry. Avicenna J. Med. Biotechnol. 6, 38–46 (2014).
Britton, S., Coates, J. & Jackson, S. P. A new method for high-resolution imaging of Ku foci to decipher mechanisms of DNA double-strand break repair. J. Cell Biol. 202, 579–595 (2013).
Marti, T. M., Hefner, E., Feeney, L., Natale, V. & Cleaver, J. E. H2AX phosphorylation within the G1 phase after UV irradiation depends on nucleotide excision repair and not DNA double-strand breaks. Proc. Natl. Acad. Sci. USA 103, 9891–9896 (2006).
Soutoglou, E. et al. Positional stability of single double-strand breaks in mammalian cells. Nat. Cell Biol. 9, 675–682 (2007).
Lan, L. et al. The ACF1 complex is required for DNA double-strand break repair in human cells. Mol. Cell 40, 976–987 (2010).
Uphoff, C. C. & Drexler, H. G. Detecting mycoplasma contamination in cell cultures by polymerase chain reaction. Methods Mol. Biol. 731, 93–103 (2011).
Janicki, S. M. et al. From silencing to gene expression: real-time analysis in single cells. Cell 116, 683–698 (2004).
Acknowledgements
The laboratory of F.d’A.d.F. was supported by the Associazione Italiana per la Ricerca sul Cancro, AIRC (application 12971), the Human Frontier Science Program (contract RGP 0014/2012), the Cariplo Foundation (grant nos. 2010-0818 and 2014-0812), Fondazione Telethon (GGP12059), the Association for International Cancer Research (AICR-Worldwide Cancer Research Rif. N. 14-1331), Progetti di Ricerca di Interesse Nazionale (PRIN) 2010/2011/2015, the Italian Ministry of Education Universities and Research EPIGEN and INTEROMICs project, the AMANDA project Accordo Quadro Regione Lombardia–CNR, AIRC Special Program ‘5 per mille’ metastases project no. 21091 and a European Research Council advanced grant (322726). S.F. was supported by Collegio Ghislieri, Fondazione Cariplo (2014-1215) and AriSLA (project ‘DDRNA&ALS’).
Author information
Authors and Affiliations
Contributions
S.F. and F.d’A.d.F. developed the initial version of the protocol. F.d’A.d.F. conceived the use of the procedure for discovering DDRNA and its functions. F.M. performed experiments with fluorescent RNA in NIH2/4 cells. F.R. adapted RATaR for Rosa26-CreERT2 Trf2F/F MEFs. S.F. adapted RATaR for U2OS-TRE19 cells. F.M. assembled the figures. F.M. and S.F. wrote the manuscript. F.R. integrated the parts relative to Rosa26-CreERT2 Trf2F/F MEFs and F.d’A.d.F. edited the manuscript.
Corresponding author
Ethics declarations
Competing interests
The autors declare no competing interests.
Additional information
Journal peer review information: Nature Protocols thanks Bernd Kaina, Kum Khanna and other anonymous reviewer(s) for their contribution to the peer review of this work.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key reference(s) using this protocol
Francia, S. et al. Nature 488, 231–235 (2012): https://doi.org/10.1038/nature11179
Francia, S. et al. J. Cell Sci. 129, 1468–1476 (2016): https://doi.org/10.1242/jcs.182188
Rossiello, F. et al. Nat. Commun. 8, 13980 (2017): https://doi.org/10.1038/ncomms13980
Michelini, F. et al. Nat. Cell Biol. 19, 1400–1411 (2017): https://doi.org/10.1038/ncb3643
Integrated supplementary information
Supplementary Figure 1 Examples of permeabilized NIH2/4 cells.
Images of non-permeabilized and permeabilized NIH2/4 cells, captured with a transmitted light microscope, show a clear difference in cell transparency and thickness. Margins of nuclei and nucleoli are more evident in permeabilized cells. As an example, the outlines of a non-permeabilized and a permeabilized cell membrane (green), nucleus (red) and nucleoli (blue) are depicted. Scale bar, 50 μM.
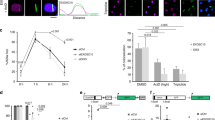
Supplementary Figure 2 RATaR shows site-specific DDRNA localization to the damage locus and 53BP1 focus reformation in U2OS TRE/I-SceI-19 cells.
U2OS TRE/I-SceI-19 cells were co-transfected with YFP-TetR- and I-SceI-expressing vectors. 24 hours later, doxycycline (1 μg/mL) was added to the medium to induce YFP-TetR binding to the TetO array, enabling visualization of the locus, and after 3 hours cells were permeabilized and treated with RNase A. Cells were then reconstituted by incubation with synthetic DDRNA-Cy5 matching TetO sequences (Tet DDRNA, 100 μM) or DDRNA-Cy5 matching LacO sequences (Lac DDRNA, 100 μM) as control, mixed with tRNA (200 ng). Images show that only DDRNAs matching TetO sequences (magenta) localize to the YFP-TetR-marked locus (green) and reconstitute 53BP1 (blue) focus post RNase A treatment, while DDRNAs matching LacO sequences do not. Images were captured by a confocal microscope. The dotted outline depicts the nucleus. Arrows indicate YFP-TetR-marked locus. Scale bar, 5 μM.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1 and 2
Rights and permissions
About this article
Cite this article
Michelini, F., Rossiello, F., d’Adda di Fagagna, F. et al. RNase A treatment and reconstitution with DNA damage response RNA in living cells as a tool to study the role of non-coding RNA in the formation of DNA damage response foci. Nat Protoc 14, 1489–1508 (2019). https://doi.org/10.1038/s41596-019-0147-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-019-0147-5
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.