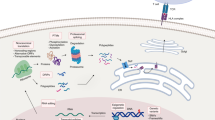
Abstract
Peptide antigens bound to molecules encoded by the major histocompatibility complex (MHC) and presented on the cell surface form the targets of T lymphocytes. This critical arm of the adaptive immune system facilitates the eradication of pathogen-infected and cancerous cells, as well as the production of antibodies. Methods to identify these peptide antigens are critical to the development of new vaccines, for which the goal is the generation of effective adaptive immune responses and long-lasting immune memory. Here, we describe a robust protocol for the identification of MHC-bound peptides from cell lines and tissues, using nano-ultra-performance liquid chromatography coupled to high-resolution mass spectrometry (nUPLC–MS/MS) and recent improvements in methods for isolation and characterization of these peptides. The protocol starts with the immunoaffinity capture of naturally processed MHC-peptide complexes. The peptides dissociate from the class I human leukocyte antigens (HLAs) upon acid denaturation. This peptide cargo is then extracted and separated into fractions by HPLC, and the peptides in these fractions are identified using nUPLC–MS/MS. With this protocol, several thousand peptides can be identified from a wide variety of cell types, including cancerous and infected cells and those from tissues, with a turnaround time of 2–3 d.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Dudek, N. L., Croft, N. P., Schittenhelm, R. B., Ramarathinam, S. H. & Purcell, A. W. A systems approach to understand antigen presentation and the immune response. Methods Mol. Biol. 1394, 189–209 (2016).
Ternette, N. et al. Early kinetics of the HLA class I-associated peptidome of MVA.HIVconsv-infected cells. J. Virol. 89, 5760–5771 (2015).
Mommen, G. P. et al. Sampling from the proteome to the human leukocyte antigen-DR (HLA-DR) ligandome proceeds via high specificity. Mol. Cell. Proteomics 15, 1412–1423 (2016).
Mommen, G. P. et al. Expanding the detectable HLA peptide repertoire using electron-transfer/higher-energy collision dissociation (EThcD). Proc. Natl Acad. Sci. USA 111, 4507–4512 (2014).
Schittenhelm, R. B., Sian, T. C., Wilmann, P. G., Dudek, N. L. & Purcell, A. W. Revisiting the arthritogenic peptide theory: quantitative not qualitative changes in the peptide repertoire of HLA-B27 allotypes. Arthritis Rheumatol. 67, 702–713 (2015).
Giam, K. et al. A comprehensive analysis of peptides presented by HLA-A1. Tissue Antigens 85, 492–496 (2015).
Caron, E. et al. An open-source computational and data resource to analyze digital maps of immunopeptidomes. Elife 4, e07661 (2015).
Bassani-Sternberg, M., Pletscher-Frankild, S., Jensen, L. J. & Mann, M. Mass spectrometry of human leukocyte antigen class I peptidomes reveals strong effects of protein abundance and turnover on antigen presentation. Mol. Cell. Proteomics 14, 658–673 (2015).
Ternette, N. et al. Defining the HLA class I-associated viral antigen repertoire from HIV-1-infected human cells. Eur. J. Immunol. 46, 60–69 (2016).
Bassani-Sternberg, M. et al. Direct identification of clinically relevant neoepitopes presented on native human melanoma tissue by mass spectrometry. Nat. Commun. 7, 13404 (2016).
Shao, W. et al. The SysteMHC Atlas project. Nucleic Acids Res. 46, D1237–D1247 (2017).
Abelin, J. G. et al. Mass spectrometry profiling of HLA-associated peptidomes in mono-allelic cells enables more accurate epitope prediction. Immunity 46, 315–326 (2017).
Khodadoust, M. S. et al. Antigen presentation profiling reveals recognition of lymphoma immunoglobulin neoantigens. Nature 543, 723–727 (2017).
Liepe, J. et al. A large fraction of HLA class I ligands are proteasome-generated spliced peptides. Science 354, 354–358 (2016).
Altman, J. D. et al. Phenotypic analysis of antigen-specific T lymphocytes. Science 274, 94–96 (1996).
McHeyzer-Williams, M. G., Altman, J. D. & Davis, M. M. Enumeration and characterization of memory cells in the TH compartment. Immunol. Rev. 150, 5–21 (1996).
Rodenko, B. et al. Generation of peptide-MHC class I complexes through UV-mediated ligand exchange. Nat. Protoc. 1, 1120–1132 (2006).
Sheikh, Q. M., Gatherer, D., Reche, P. A. & Flower, D. R. Towards the knowledge-based design of universal influenza epitope ensemble vaccines. Bioinformatics 32, 3233-3239 (2016).
Clemens, E. B. et al. Towards identification of immune and genetic correlates of severe influenza disease in indigenous Australians. Immunol. Cell Biol. 94, 367–377 (2016).
Assarsson, E. et al. Immunomic analysis of the repertoire of T-cell specificities for influenza A virus in humans. J. Virol. 82, 12241–12251 (2008).
Parra-Lopez, C. et al. Major histocompatibility complex and T cell interactions of a universal T cell epitope from Plasmodium falciparum circumsporozoite protein. J. Biol. Chem. 281, 14907–14917 (2006).
Nardin, E. H. et al. A totally synthetic polyoxime malaria vaccine containing Plasmodium falciparum B cell and universal T cell epitopes elicits immune responses in volunteers of diverse HLA types. J. Immunol. 166, 481–489 (2001).
Purcell, A. W., McCluskey, J. & Rossjohn, J. More than one reason to rethink the use of peptides in vaccine design. Nat. Rev. Drug Discov. 6, 404–414 (2007).
Brennick, C. A., George, M. M., Corwin, W. L., Srivastava, P. K. & Ebrahimi-Nik, H. Neoepitopes as cancer immunotherapy targets: key challenges and opportunities. Immunotherapy 9, 361–371 (2017).
Verdegaal, E. M. E. & van der Burg, S. H. The potential and challenges of exploiting the vast but dynamic neoepitope landscape for immunotherapy. Front. Immunol. 8, 1113 (2017).
Nepom, G. T. MHC class II tetramers. J. Immunol. 188, 2477–2482 (2012).
Crawford, F. et al. Specificity and detection of insulin-reactive CD4+ T cells in type 1 diabetes in the nonobese diabetic (NOD) mouse. Proc. Natl Acad. Sci. USA 108, 16729–16734 (2011).
Coles, R. M. et al. Virus infection expands a biased subset of T cells that bind tetrameric class I peptide complexes. Eur. J. Immunol. 33, 1557–1567 (2003).
Gray, C. M. et al. Frequency of class I HLA-restricted anti-HIV CD8+ T cells in individuals receiving highly active antiretroviral therapy (HAART). J. Immunol. 162, 1780–1788 (1999).
Bieganowska, K. et al. Direct analysis of viral-specific CD8+ T cells with soluble HLA-A2/Tax11-19 tetramer complexes in patients with human T cell lymphotropic virus-associated myelopathy. J. Immunol. 162, 1765–1771 (1999).
Murali-Krishna, K. et al. Counting antigen-specific CD8 T cells: a reevaluation of bystander activation during viral infection. Immunity 8, 177–187 (1998).
Reguzova, A. Y., Karpenko, L. I., Mechetina, L. V. & Belyakov, I. M. Peptide-MHC multimer-based monitoring of CD8 T-cells in HIV-1 infection and AIDS vaccine development. Expert Rev. Vaccines 14, 69–84 (2015).
Bacher, P. & Scheffold, A. Flow-cytometric analysis of rare antigen-specific T cells. Cytometry A 83, 692–701 (2013).
Gojanovich, G. S. et al. The use of peptide-major-histocompatibility-complex multimers in type 1 diabetes mellitus. J. Diabetes Sci. Technol. 6, 515–524 (2012).
Castelli, C. et al. Mass spectrometric identification of a naturally processed melanoma peptide recognized by CD8+ cytotoxic T lymphocytes. J. Exp. Med. 181, 363–368 (1995).
Storkus, W. J., Zeh, H. J., Salter, R. D. & Lotze, M. T. Identification of T-cell epitopes: rapid isolation of class I-presented peptides from viable cells by mild acid elution. J. Immunother. 14, 94–103 (1993).
Storkus, W. J., Zeh, H. J., Maeurer, M. J., Salter, R. D. & Lotze, M. T. Identification of human melanoma peptides recognized by class I restricted tumor infiltrating T lymphocytes. J. Immunol. 151, 3719–3727 (1993).
Barnea, E. et al. Analysis of endogenous peptides bound by soluble MHC class I molecules: a novel approach for identifying tumor-specific antigens. Eur. J. Immunol. 32, 213–222 (2002).
Buchsbaum, S. et al. Large-scale analysis of HLA peptides presented by HLA-Cw4. Immunogenetics 55, 172–176 (2003).
Wahl, A. et al. HLA class I molecules consistently present internal influenza epitopes. Proc. Natl Acad. Sci. USA 106, 540–545 (2009).
Bassani-Sternberg, M. et al. Soluble plasma HLA peptidome as a potential source for cancer biomarkers. Proc. Natl Acad. Sci. USA 107, 18769–18776 (2010).
Scull, K. E. et al. Secreted HLA recapitulates the immunopeptidome and allows in-depth coverage of HLA A*02:01 ligands. Mol. Immunol. 51, 136–142 (2012).
Yaciuk, J. C. et al. Direct interrogation of viral peptides presented by the class I HLA of HIV-infected T cells. J. Virol. 88, 12992–13004 (2014).
Norcross, M. A. et al. Abacavir induces loading of novel self-peptides into HLA-B*57: 01: an autoimmune model for HLA-associated drug hypersensitivity. Aids 26, F21–F29 (2012).
Trolle, T. et al. The length distribution of class I–restricted T cell epitopes is determined by both peptide supply and MHC allele–specific binding preference. J. Immunol. 196, 1480–1487 (2016).
Kaabinejadian, S. et al. Immunodominant West Nile virus T cell epitopes are fewer in number and fashionably late. J. Immunol. 196, 4263–4273 (2016).
Abelin, J. G. et al. Complementary IMAC enrichment methods for HLA-associated phosphopeptide identification by mass spectrometry. Nat. Protoc. 10, 1308–1318 (2015).
Gilchuk, P. et al. Discovering naturally processed antigenic determinants that confer protective T cell immunity. J. Clin. Invest. 123, 1976–1987 (2013).
Corr, M. et al. Endogenous peptides of a soluble major histocompatibility complex class I molecule, H-2Lds: sequence motif, quantitative binding, and molecular modeling of the complex. J. Exp. Med. 176, 1681–1692 (1992).
Joyce, S. & Nathenson, S. G. Methods to study peptides associated with MHC class I molecules. Curr. Opin. Immunol. 6, 24–31 (1994).
Hunt, D. F. et al. Characterization of peptides bound to the class I MHC molecule HLA-A2.1 by mass spectrometry. Science 255, 1261–1263 (1992).
Engelhard, V. H. et al. Mass spectrometric analysis of peptides associated with the human class I MHC molecules HLA-A2.1 and HLA-B7 and identification of structural features that determine binding. Chem. Immunol. 57, 39–62 (1993).
Cox, A. L. et al. Identification of a peptide recognized by five melanoma-specific human cytotoxic T cell lines. Science 264, 716–719 (1994).
Slingluff, C. L. Jr, Hunt, D. F. & Engelhard, V. H. Direct analysis of tumor-associated peptide antigens. Curr. Opin. Immunol. 6, 733–740 (1994).
Skipper, J. C. et al. An HLA-A2-restricted tyrosinase antigen on melanoma cells results from posttranslational modification and suggests a novel pathway for processing of membrane proteins. J. Exp. Med. 183, 527–534 (1996).
Meadows, L. et al. The HLA-A*0201-restricted H-Y antigen contains a posttranslationally modified cysteine that significantly affects T cell recognition. Immunity 6, 273–281 (1997).
Crotzer, V. L. et al. Immunodominance among EBV-derived epitopes restricted by HLA-B27 does not correlate with epitope abundance in EBV-transformed B-lymphoblastoid cell lines. J. Immunol. 164, 6120–6129 (2000).
Zarling, A. L. et al. Phosphorylated peptides are naturally processed and presented by major histocompatibility complex class I molecules in vivo. J. Exp. Med. 192, 1755–1762 (2000).
Mohammed, F. et al. Phosphorylation-dependent interaction between antigenic peptides and MHC class I: a molecular basis for the presentation of transformed self. Nat. Immunol. 9, 1236–1243 (2008).
Van Bleek, G. M. & Nathenson, S. G. Isolation of an endogenously processed immunodominant viral peptide from the class I H-2Kb molecule. Nature 348, 213–216 (1990).
Rotzschke, O. et al. Isolation and analysis of naturally processed viral peptides as recognized by cytotoxic T cells. Nature 348, 252–254 (1990).
Falk, K. et al. Identification of naturally processed viral nonapeptides allows their quantification in infected cells and suggests an allele-specific T cell epitope forecast. J. Exp. Med. 174, 425–434 (1991).
Falk, K., Rötzschke, O., Stevanovic, S., Jung, G. & Rammensee, H.-G. Allele-specific motifs revealed by sequencing of self-peptides eluted from MHC molecules. Nature 351, 290–296 (1991).
Falk, K., Rötzschke, O. & Rammensee, H.-G. A self peptide naturally presented by both H-2Kb and H-2Kbm1 molecules demonstrates MHC restriction of self tolerance at the molecular level. Int. Immunol. 4, 321–325 (1992).
Falk, K., Rotzschke, O., Stevanovic, S., Jung, G. & Rammensee, H. G. Pool sequencing of natural HLA-DR, DQ, and DP ligands reveals detailed peptide motifs, constraints of processing, and general rules. Immunogenetics 39, 230–242 (1994).
Rammensee, H.-G. Chemistry of peptide associated with class I and class II molecules. Curr. Opin. Immunol. 7, 85–95 (1995).
Rammensee, H., Bachmann, J., Emmerich, N. P., Bachor, O. A. & Stevanovic, S. SYFPEITHI: database for MHC ligands and peptide motifs. Immunogenetics 50, 213–219 (1999).
Schittenhelm, R. B., Sivaneswaran, S., Lim Kam Sian, T. C., Croft, N. P. & Purcell, A. W. Human leukocyte antigen (HLA) B27 allotype-specific binding and candidate arthritogenic peptides revealed through heuristic clustering of data-independent acquisition mass spectrometry (DIA-MS) data. Mol. Cell. Proteomics 15, 1867–1876 (2016).
Trujillo, J. A. et al. The cellular redox environment alters antigen presentation. J. Biol. Chem. 289, 27979–27991 (2014).
Scally, S. W. et al. A molecular basis for the association of the HLA-DRB1 locus, citrullination, and rheumatoid arthritis. J. Exp. Med. 210, 2569–2582 (2013).
Illing, P. T. et al. Immune self-reactivity triggered by drug-modified HLA-peptide repertoire. Nature 486, 554–558 (2012).
Wynne, J. W. et al. Characterization of the antigen processing machinery and endogenous peptide presentation of a bat MHC class I molecule. J. Immunol. 196, 4468–4476 (2016).
Edwards, P. A., Smith, C. M., Neville, A. M. & O’Hare, M. J. A human-hybridoma system based on a fast-growing mutant of the ARH-77 plasma cell leukemia-derived line. Eur. J. Immunol. 12, 641–648 (1982).
Alexander, J., Payne, J. A., Murray, R., Frelinger, J. A. & Cresswell, P. Differential transport requirements of HLA and H-2 class I glycoproteins. Immunogenetics 29, 380–388 (1989).
Kavathas, P., Bach, F. H. & DeMars, R. Gamma ray-induced loss of expression of HLA and glyoxalase I alleles in lymphoblastoid cells. Proc. Natl Acad. Sci. USA 77, 4251–4255 (1980).
Shimizu, Y., Geraghty, D. E., Koller, B. H., Orr, H. T. & DeMars, R. Transfer and expression of three cloned human non-HLA-A,B,C class I major histocompatibility complex genes in mutant lymphoblastoid cells. Proc. Natl Acad. Sci. USA 85, 227–231 (1988).
Partridge, T. et al. Discrimination between human leukocyte antigen class I–bound and co-purified HIV-derived peptides in immunopeptidomics workflows. Front. Immunol. 9, 912 (2018).
Riberdy, J. M., Newcomb, J. R., Surman, M. J., Barbosa, J. A. & Cresswell, P. HLA-DR molecules from an antigen-processing mutant cell line are associated with invariant chain peptides. Nature 360, 474–477 (1992).
Nelson, C. A., Roof, R. W., McCourt, D. W. & Unanue, E. R. Identification of the naturally processed form of hen egg white lysozyme bound to the murine major histocompatibility complex class II molecule I-Ak. Proc. Natl Acad. Sci. USA 89, 7380–7383 (1992).
Perkins, D. N., Pappin, D. J., Creasy, D. M. & Cottrell, J. S. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20, 3551–3567 (1999).
Eng, J. K., McCormack, A. L. & Yates, J. R. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 5, 976–989 (1994).
Craig, R. & Beavis, R. C. TANDEM: matching proteins with tandem mass spectra. Bioinformatics 20, 1466–1467 (2004).
Tyanova, S., Temu, T. & Cox, J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 11, 2301–2319 (2016).
Shilov, I. V. et al. The Paragon Algorithm, a next generation search engine that uses sequence temperature values and feature probabilities to identify peptides from tandem mass spectra. Mol. Cell. Proteomics 6, 1638–1655 (2007).
Zhang, J. et al. PEAKS DB: de novo sequencing assisted database search for sensitive and accurate peptide identification. Mol. Cell. Proteomics 11, M111.010587 (2012).
Giansanti, P., Tsiatsiani, L., Low, T. Y. & Heck, A. J. Six alternative proteases for mass spectrometry–based proteomics beyond trypsin. Nat. Protoc. 11, 993–1006 (2016).
Meyer, J. G. et al. Expanding proteome coverage with orthogonal-specificity alpha-lytic proteases. Mol. Cell. Proteomics 13, 823–835 (2014).
Faridi, P., Purcell, A. W. & Croft, N. P. In immunopeptidomics we need a sniper instead of a shotgun. Proteomics 18, e1700464 (2018).
Nielsen, M. et al. Reliable prediction of T-cell epitopes using neural networks with novel sequence representations. Protein. Sci. 12, 1007–1017 (2003).
Andreatta, M. & Nielsen, M. Gapped sequence alignment using artificial neural networks: application to the MHC class I system. Bioinformatics 32, 511–517 (2016).
Schneider, T. D. & Stephens, R. M. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 18, 6097–6100 (1990).
Thomsen, M. C. F. & Nielsen, M. Seq2Logo: a method for construction and visualization of amino acid binding motifs and sequence profiles including sequence weighting, pseudo counts and two-sided representation of amino acid enrichment and depletion. Nucleic Acids Res. 40, W281–W287 (2012).
Andreatta, M., Lund, O. & Nielsen, M. Simultaneous alignment and clustering of peptide data using a Gibbs sampling approach. Bioinformatics 29, 8–14 (2013).
Bassani-Sternberg, M. et al. Deciphering HLA-I motifs across HLA peptidomes improves neo-antigen predictions and identifies allostery regulating HLA specificity. PLoS Comput. Biol. 13, e1005725 (2017).
Nepom, B. S., Nepom, G. T., Coleman, M. & Kwok, W. W. Critical contribution of beta chain residue 57 in peptide binding ability of both HLA-DR and -DQ molecules. Proc. Natl Acad. Sci. USA 93, 7202–7206 (1996).
Lampson, L. A. & Levy, R. Two populations of Ia-like molecules on a human B cell line. J. Immunol. 125, 293–299 (1980).
Gorga, J. C., Knudsen, P. J., Foran, J. A., Strominger, J. L. & Burakoff, S. J. Immunochemically purified DR antigens in liposomes stimulate xenogeneic cytolytic T cells in secondary in vitro cultures. Cell. Immunol. 103, 160–173 (1986).
Kolstad, A., Johansen, B. & Hannestad, K. Two HLA-DQ-specific human-human hybridoma antibodies (TrG6;TrC5) define epitopes also expressed by a transcomplementing hybrid DQ molecule (DQw7 alpha/DQw4 beta). Hum. Immunol. 24, 15–29 (1989).
Watson, A. J., DeMars, R., Trowbridge, I. S. & Bach, F. H. Detection of a novel human class II HLA antigen. Nature 304, 358–361 (1983).
Ellis, S. A., Taylor, C. & McMichael, A. Recognition of HLA-B27 and related antigen by a monoclonal antibody. Hum. Immunol. 5, 49–59 (1982).
Parham, P. & Brodsky, F. M. Partial purification and some properties of BB7.2. A cytotoxic monoclonal antibody with specificity for HLA-A2 and a variant of HLA-A28. Hum. Immunol. 3, 277–299 (1981).
Schittenhelm, R. B., Dudek, N. L., Croft, N. P., Ramarathinam, S. H. & Purcell, A. W. A comprehensive analysis of constitutive naturally processed and presented HLA-C*04:01 (Cw4)-specific peptides. Tissue Antigens 83, 174–179 (2014).
Braud, V. M., Allan, D. S., Wilson, D. & McMichael, A. J. TAP- and tapasin-dependent HLA-E surface expression correlates with the binding of an MHC class I leader peptide. Curr. Biol. 8, 1–10 (1998).
Thomas, R. et al. HLA-C cell surface expression and control of HIV/AIDS correlate with a variant upstream of HLA-C. Nat. Genet. 41, 1290–1294 (2009).
Acknowledgements
We thank the following funding sources for supporting this work: Australian National Health and Medical Research Council (NHMRC) project grants APP1085018 and APP1084283, and Australian Research Council (ARC) project grant DP150104503. A.W.P. was supported by an NHMRC Principal Research Fellowship (APP 1137739).
Author information
Authors and Affiliations
Contributions
A.W.P., S.H.R. and N.T. wrote the manuscript and collated data and figures.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key reference(s) using this protocol
Schittenhelm, R. B., Dudek, N. L., Croft, N. P., Ramarathinam, S. H. & Purcell, A. W. Mol. Cell. Proteomics 15, 1867–1876 (2016): https://doi.org/10.1074/mcp.M115.056358
Scally, S. W. et al. J. Exp. Med. 210, 2569–2582 (2013): https://doi.org/10.1084/jem.20131241
Illing, P. T. et al. Nature 486, 554–558 (2012): https://doi.org/10.1038/nature11147
Wynne, J. W. et al. J. Immunol. 196, 4468–4476 (2016): https://doi.org/10.4049/jimmunol.1502062
Rights and permissions
About this article
Cite this article
Purcell, A.W., Ramarathinam, S.H. & Ternette, N. Mass spectrometry–based identification of MHC-bound peptides for immunopeptidomics. Nat Protoc 14, 1687–1707 (2019). https://doi.org/10.1038/s41596-019-0133-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-019-0133-y
This article is cited by
-
Immunopeptidomics-based identification of naturally presented non-canonical circRNA-derived peptides
Nature Communications (2024)
-
Thunder-DDA-PASEF enables high-coverage immunopeptidomics and is boosted by MS2Rescore with MS2PIP timsTOF fragmentation prediction model
Nature Communications (2024)
-
Protein degradation by human 20S proteasomes elucidates the interplay between peptide hydrolysis and splicing
Nature Communications (2024)
-
Antigen identification strategies and preclinical evaluation models for advancing tuberculosis vaccine development
npj Vaccines (2024)
-
Adoptive neoantigen-reactive T cell therapy: improvement strategies and current clinical researches
Biomarker Research (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.