Abstract
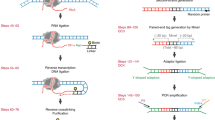
Alterations in chromatin structure play a major role in the epigenetic regulation of gene expression. Here, we describe a step-by-step protocol for differential viral accessibility (DIVA), a method for identifying changes in chromatin accessibility genome-wide. Commonly used methods for mapping accessible genomic loci have strong preferences toward detecting ‘open’ chromatin found at regulatory regions but are not well suited to studying chromatin accessibility in gene bodies and intergenic regions. DIVA overcomes this limitation, enabling a broader range of sites to be interrogated. Conceptually, DIVA is similar to ATAC-seq in that it relies on the integration of exogenous DNA into the genome to map accessible chromatin, except that chromatin architecture is probed through mapping integration sites of exogenous lentiviruses. An isogenic pair of cell lines are transduced with a lentiviral vector, followed by PCR amplification and Illumina sequencing of virus–genome junctions; the resulting sequences define a set of unique lentiviral integration sites, which are compared to determine whether genomic loci exhibit significantly altered accessibility between experimental and control cells. Experienced researchers will take 6 d to generate lentiviral stocks and transduce the target cells, a further 5 d to prepare the Illumina sequencing libraries and a few hours to perform the bioinformatic analysis.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Boyle, A. P. et al. High-resolution mapping and characterization of open chromatin across the genome. Cell 132, 311–322 (2008).
Schones, D. E. et al. Dynamic regulation of nucleosome positioning in the human genome. Cell 132, 887–898 (2008).
Giresi, P. G., Kim, J., McDaniell, R. M., Iyer, V. R. & Lieb, J. D. FAIRE (formaldehyde-assisted isolation of regulatory elements) isolates active regulatory elements from human chromatin. Genome Res. 17, 877–885 (2007).
Gargiulo, G. et al. NA-Seq: a discovery tool for the analysis of chromatin structure and dynamics during differentiation. Dev. Cell 16, 466–481 (2009).
Chen, P. B., Zhu, L. J., Hainer, S. J., McCannell, K. N. & Fazzio, T. G. Unbiased chromatin accessibility profiling by RED-seq uncovers unique features of nucleosome variants in vivo. BMC Genomics 15, 1104 (2014).
Tsompana, M. & Buck, M. J. Chromatin accessibility: a window into the genome. Epigenetics Chromatin 7, 33 (2014).
Tchasovnikarova, I. A. et al. Epigenetic silencing by the HUSH complex mediates position-effect variegation in human cells. Science 348, 1481–1485 (2015).
Timms, R. T., Tchasovnikarova, I. A. & Lehner, P. J. Position-effect variegation revisited: HUSHing up heterochromatin in human cells. Bioessays 38, 333–343 (2016).
Tchasovnikarova, I. A. et al. Hyperactivation of HUSH complex function by Charcot–Marie–Tooth disease mutation in MORC2. Nat. Genet. 49, 1035–1044 (2017).
Timms, R. T., Tchasovnikarova, I. A., Antrobus, R., Dougan, G. & Lehner, P. J. ATF7IP-mediated stabilization of the histone methyltransferase SETDB1 is essential for heterochromatin formation by the HUSH complex. Cell Rep. 17, 653–659 (2016).
Buenrostro, J. D., Giresi, P. G., Zaba, L. C., Chang, H. Y. & Greenleaf, W. J. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218 (2013).
Kvaratskhelia, M., Sharma, A., Larue, R. C., Serrao, E. & Engelman, A. Molecular mechanisms of retroviral integration site selection. Nucleic Acids Res. 42, 10209–10225 (2014).
Buenrostro, J. D. et al. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature 523, 486–490 (2015).
Debyser, Z., Christ, F., De Rijck, J. & Gijsbers, R. Host factors for retroviral integration site selection. Trends Biochem. Sci. 40, 108–116 (2015).
Carette, J. E. et al. Global gene disruption in human cells to assign genes to phenotypes by deep sequencing. Nat. Biotechnol. 29, 542–546 (2011).
Jae, L. T. et al. Virus entry. Lassa virus entry requires a trigger-induced receptor switch. Science 344, 1506–1510 (2014).
Blomen, V. A. et al. Gene essentiality and synthetic lethality in haploid human cells. Science 350, 1092–1096 (2015).
Timms, R. T. et al. Genetic dissection of mammalian ERAD through comparative haploid and CRISPR forward genetic screens. Nat. Commun. 7, 11786 (2016).
Quinlan, A. R. & Hall, I. M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010).
Sambrook, J. & Russell, D. W. Purification of nucleic acids by extraction with phenol:chloroform. CSH Protoc. 2006, https://doi.org/10.1101/pdb.prot4455 (2006).
Kundaje, A. et al. Integrative analysis of 111 reference human epigenomes. Nature 518, 317–330 (2015).
Acknowledgements
We thank S. Andrews for assistance with data analysis using SeqMonk. This work was supported by the Wellcome Trust, through a Principal Research Fellowship to P.J.L. (101835/Z/13/Z), a Sir Henry Wellcome Fellowship to R.T.T. (201387/Z/16/Z) and a PhD studentship to I.A.T. I.A.T. is a Damon Runyon Fellow supported by the Damon Runyon Cancer Research Foundation (DRG-2277-16).
Author information
Authors and Affiliations
Contributions
R.T.T., I.A.T. and P.J.L. conceived the method. I.A.T. and R.T.T. performed all the experiments and, together with P.J.L., analyzed the data and wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related link
Key reference using this protocol
Tchasovnikarova, I. A. et al. Nat. Genet. 49, 1035–1044 (2017): https://doi.org/10.1038/ng.3878
Rights and permissions
About this article
Cite this article
Timms, R.T., Tchasovnikarova, I.A. & Lehner, P.J. Differential viral accessibility (DIVA) identifies alterations in chromatin architecture through large-scale mapping of lentiviral integration sites. Nat Protoc 14, 153–170 (2019). https://doi.org/10.1038/s41596-018-0087-5
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-018-0087-5
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.